|
|
[ Download Map Data ]
[ Download Feature Correspondence Data ]
|
|
|
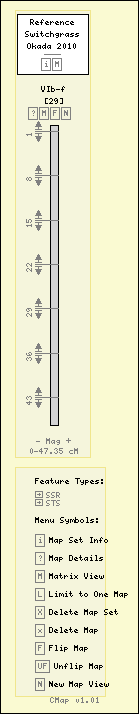
Switchgrass - Okada 2010 - VIb-f
(Click headers to resort) |
Comparative Maps |
| Feature |
Type |
Position (cM) |
Map |
Feature |
Type |
Position |
Evidence |
Actions |
|
sww1926_220
|
STS
|
44.24
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g026520
|
Gene Prediction
|
61,665,164 - 61,666,734 ()
|
related_sequence
|
View Maps
|
|
sww3033_180
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww3033_186
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww381_217
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww381_219
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww2284_202
|
STS
|
6.13
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g001130
|
Gene Prediction
|
1,101,094 - 1,106,395 ()
|
related_sequence
|
View Maps
|
|
sww2580_204
|
STS
|
2.19
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g000625
|
Gene Prediction
|
448,614 - 448,937 ()
|
related_sequence
|
View Maps
|
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 1
|
Sb01g018170
|
Gene Prediction
|
18,874,567 - 18,874,848 ()
|
related_sequence
|
View Maps
|
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 1
|
Sb01g009730
|
Gene Prediction
|
8,502,005 - 8,503,090 ()
|
related_sequence
|
View Maps
|
|
sww1889_222
|
STS
|
0
|
No other positions |
|
sww3026_160
|
STS
|
47.35
|
No other positions |
|
sww47_249
|
STS
|
45.55
|
No other positions |
|
sww1926_220
|
STS
|
44.24
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g026520
|
Gene Prediction
|
61,665,164 - 61,666,734 ()
|
related_sequence
|
View Maps
|
|
sww1573_225
|
STS
|
33.07
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g022240
|
Gene Prediction
|
56,673,459 - 56,675,165 ()
|
related_sequence
|
View Maps
|
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 2
|
Sb02g025640
|
Gene Prediction
|
60,605,341 - 60,606,234 ()
|
related_sequence
|
View Maps
|
|
sww1549_210
|
STS
|
27.37
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g022240
|
Gene Prediction
|
56,673,459 - 56,675,165 ()
|
related_sequence
|
View Maps
|
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 2
|
Sb02g025640
|
Gene Prediction
|
60,605,341 - 60,606,234 ()
|
related_sequence
|
View Maps
|
|
nfsg057_121
|
SSR
|
23.56
|
No other positions |
|
nfsg057_126
|
SSR
|
23.56
|
No other positions |
|
sww3043_168
|
STS
|
21.48
|
No other positions |
|
sww3043_170
|
STS
|
21.48
|
No other positions |
|
sww2180_359
|
STS
|
19.85
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g003440
|
Gene Prediction
|
4,139,923 - 4,142,365 ()
|
related_sequence
|
View Maps
|
|
sww3046_178
|
STS
|
19.85
|
No other positions |
|
sww3046_182
|
STS
|
19.85
|
No other positions |
|
sww3033_180
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww3033_186
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww381_217
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww381_219
|
STS
|
18.27
|
Sorghum - Sorghum bicolor JGI8X 2007 - Chr. 7
|
Sb07g002950
|
Gene Prediction
|
3,115,804 - 3,117,304 ()
|
related_sequence
|
View Maps
|
|
sww3039_165
|
STS
|
16
|
No other positions |