|
Over 22 bacterial blight (BB) resistance genes have been identified from
cultivated rice and its wild relatives (Khush and Angeles 1999). The recessive
gene xa8 was originally identified from a rice line PI 231129 (Sidhu
et al., 1978). It showed resistance to five out of six races of
the pathogen Xanthomonas oryzae pv oryzae (Xoo) in
the Philippines (Khush, 1992). The gene was transferred into IR24 through
repeated backcrossing resulting in the production of a near-isogenic line
IRBB8 carrying xa8 in IR24 genetic background (Ogawa et al.,
1991). The xa8 gene is of special interest to breeding programs
in India as it confers moderate to high level of resistance to most of
the prevalent Xoo isolates in the Punjab province of India (unpublished
results). So far, the chromosomal location of xa8 is not known.
Identification of linked markers would be particularly useful in tracking
the incorporation of the recessive gene into advanced breeding lines.
As a first step, we here report mapping of xa8 using sequence tagged
microsatellite (STMS) markers.
A set of 105 F5 recombinant inbred lines (RILs), generated
by single seed descent from a cross of PR106 with IRBB8, was used as the
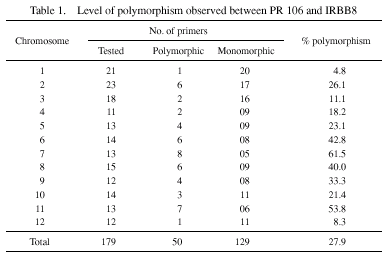
mapping population. A total of 179 STMS markers covering 12 linkage groups
were used for polymorphism survey of the parental lines, PR 106 and IRBB8
(Temnykh et al., 2000). The parental lines PR 106 and IRBB8 have
a common parent (IR8) in their pedigrees, and hence the overall level
of polymorphism between the parental lines was relatively low (27.9%)
except for chromosomes 7 and 11 which showed 61.5% and 53.8% polymorphism,
respectively (Table 1). Interestingly, STMS markers on a 33-cM region
of chromosomes 7 and a 46-cM region of chromosome 11 were all polymorphic,
suggesting that these two regions might have been introgressed from the
donor line PI231129.
All 105 F5 RILs together with the parents were analyzed using
polymorphic STMS markers on chromosomes 7 and 11. DNA was isolated from
six plants in each RIL using the CTAB method (Saghai-Maroof et al.,
1984). PCR reactions were performed as described by Chen et al.
(1997). The PCR products were separated on 4% polyacrylamide denaturing
gels and marker bands were visualized using the silver staining protocol
(Panaud et al., 1996). Banding pattern of each line was recorded
as PR 106 type or IRBB8 type or heterozygous.
The parents and 105 RILs were inoculated with four Philippine races (PXO
86, PXO 112, PXO 340 and PXO 341) and four races from Punjab (designated
I, II, III, IV) (G. L. Raina - personal communication). Six plants in
each RIL (67-day after seeding) were inoculated with each of the strains.
Lesion length was recorded on three leaves of each plant 14 days after
inoculation. Average lesion length for each plant was calculated and the
RILs were classified as IRBB8 type or PR106 type. RILs that showed variation
in disease score within a line were


classified as segregating. The RILs showing lesion length more than 10
cm against PXO 340 and PXO 341 were classified as susceptible. IRBB8 showed
partial resistance to PXO 86 and PXO 112, whereas PR106 showed susceptible
reaction to all the four Philippine races (Table 2). IRBB8 showed partial
resistance to races I, II, and IV and complete resistance to race III
in Punjab (Table 2).
Segregation analysis was conducted by inoculating the RILs with Punjab
race III. Out of a total 105 RILs, 51 showed IRBB8 type lesion length,
47 were PR 106 type and 7 were segregating. The segregation pattern was
consistent with the expected segregation ratio for a single recessive
gene in F5 ( chi2 = 0.86). Linkage analysis using
Mapmaker (Mapmaker/Exp. V3.0.) placed xa8 19.9 cM from microsatellite
marker RM214 on chromosome 7 (Fig. 1). No polymorphic markers have yet
been found on the other side of xa8. For effective marker assisted
selection, more closely linked markers flanking both sides of xa8
are needed.
Help provided by Dr G. L. Raina, PAU, RRS Kapurthala, for screening the
RILs against the Punjab strains is gratefully acknowledged.
References
Chen, X, S. Temnykh, Y. Xu, Y. G. Cho, and S. R. McCouch. 1997. Development
of a microsatellite framework map providing genome-wide coverage in rice
(Oryza sativa L.). Theor. Appl. Genet. 95: 553-567.
Khush, G. S. 1992 Selecting rice for simply inherited resistance. In:
H.T.Stalker and J.P.Murphy (eds), Plant Breeding in 1990s. CAB International,
Wallingford, UK. p. 303-322.
Khush, G. S., E. R. Angeles. 1999. A new gene for resistance to race 6
of bacterial blight in rice Oryza sativa. RGN 16: 92-93.
Ogawa, T., Y. Yamamoto, G. S. Khush and T. W. Mew. 1991. Breeding near-isogenic
lines of rice with single gene for resistance to bacterial blight pathogen
(Xanthomonas campestris pv oryzae). Jpn. J. Breed. 41: 523-529.
Panaud, O., X. Chen, and S. R. McCouch. 1996. Development of microsatellite
marker sand characterization of simple sequence length polymorphism (SSLP)
in rice (Oryza sativa L.). Mol. Gen. Genet. 252: 597-607.
Saghai-Maroof, M. A., K. M. Soliman. R. A. Jorgensen and R. W. Allard.
1984. Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance,
chromosomal locations and population dynamics. Proc. Natl. Acad. Sci.
USA 81: 8014-8018.
Sidhu G. S., G. S. Khush and T. W. Mew. 1978. Genetic analysis of bacterial
blight resistance to seventy-four cultivars of rice Oryza sativa L.
Theor. Appl. Genet. 53: 105-111.
Temnykh, S., W. D. Park, N. Ayres, S. Cartinhour, N. Hauck, L. Lipovich,
Y. G. Cho, T. Ishii and S. R. McCouch. 2000. Mapping and genome organization
of microsatellite sequences in rice (Oryza sativa L.). Theor. Appl.
Genet. 100: 697-712.
|