derivatives
EM. ABBASI’, D.S. Brar1, A.L. CARPENA2 and G.S. Khush1
The genus Oryza is comprised of 24 species, having 2n = 24
or 48 chromosomes representing nine different genomes; AA, BB, CC, BBCC,
CCDD, EE, FF, GG and HHJJ genomes. These have been classified into four
species complexes sativa, officinalis, ridleyi, and meyeriana. However,
0. brachyantha, an African species is not included in any of these complexes
(Vaughan 1989). Oryza brachyantha (2n = 24, FF genome) possesses useful
genes for resistance to stemborer, leaffolder, bacterial blight, blast
and whorl maggot. A series of hybrids have been produced through direct
crosses and embryo rescue between elite breeding lines of rice and wild
species, representing each of the nine genomes (Brar and Khush 1995). In
addition, monosomic alien addition lines (MAALs) 2n = 25 and introgression
lines have been produced from several cross-combinations of cultivated
and wild species.
We used genomic in situ hybridization (GISH) to characterize
parental chromosomes in the F1 hybrid of 0. sativa cv 1R56 and 0. brachyantha
(accession 101232) and to identify extra chromosome in monosomic alien
addition lines. The in situ hybridization protocol as given by Rayburn
and Gill (1985) was followed with minor modifications. Total genomic DNA
of 0. brachyantha was digested with EcoRI and labelled with biotin-l4-
dATP and used as a probe. Somatic chromosome analysis showed 2n =24 in
both parental species in 0. sativa and 0. brachyantha and 24 chromosomes
in the F1 hybrid. Meiotic analysis showed regular formation of 12 at metaphase
I in both the parents. On the other hand, the Fl hybrid showed limited
chromosome pairing (0- 2 bivalents), between sativa and brachyantha chromosomes,
average being 0.05 bivalents per cell. In majority of the cells, the Fl
showed unpaired chromosomes as 24 univalents. At anaphase, unequal distribution
of chromosomes was commonly observed.
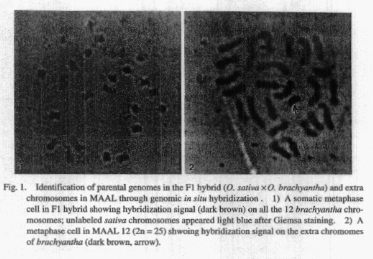
After in situ hybridization, all the 12 chromosomes of 0.
brachyantha could be identified from sativa chromosomes. The hybridization
signal appeared dark brown due to probe hybridization and non labelled
sativa chromosomes appeared light blue (Fig. 1-
1). In a similar experiment, two MAALs (MAAL 6 and MAAL 12)
derived from 0. brachyantha were used for in situ hybridization. The extra
chromosome of 0. brachyantha
showed dark brown hybridization signal (arrow in Fig. 1-2)
making it clearly distinguishable from the remaining 24 chromosomes of
0. sativa.
The results show that there is very limited homoeology between
chromosomes of 0. sativa and 0. brachyantha. Aggarwal et al. (1977) also
demonstrated through total gemonic DNA hybridization that the genomes of
0. sativa and 0. brachyantha are highly diverged. The chromosomes of parental
species in Fl hybrid and extra chromosome in MAAL could be easily distinguished
through GISH. The technique is being used to characterize introgression
of alien chromosome segments from brachyantha into the sativa genome.
Aggarwal, R.K., D.S. Brar and G.S. Khush, 1997. Two new genomes
in the Oryza complex identified on the basis of molecular divergence analysis
using total genomic DNA hybridization. Mol. Gen. Genet. 252:
1-12.
Brar, D.S. and G.S. Khush, 1995. Wide hybridization for enhancing
resistance to biotic and abiotic stresses in rainfed lowland rice. Pages
901-910. In: Proc. Intern. Rice Research Conference, IRRI, Manila, Philippines.
Rayburn, A.L. and B.S. Gill, 1985. Use of biotin-labeled
probes to map specific DNA sequences on wheat chromosomes. J. Hered. 76:
76-81.
Vaughan, D.A., 1989. The genus Oryza L. current status of
taxonomy. IRRI Research Paper Series No. 138. pp.21.
|