II. Cytogenetics
officinalis Wall ex Watt. through fluorescence in situ hybridization
M. ASGHAR’, D.S. Brar’, J.E. Hernandez2, N. Ohmido3 and G.S.
KHUSH1
Flourescence in situ hybridization (FISH) is a useful method
for characterization of parental genomes in wide hybrids, detection of
alien introgression and in phylogenetic analysis. We applied FISH to identify
parental chromosomes and to locate rDNA loci in F1 hybrid of an elite breeding
line of Oryza sativa and 0. officinalis (accession 100896). Somatic chromosome
preparations of parents and F1 hybrid were made using enzymatic maceration
method. Total genomic DNA of 0. officinalis and 45S rDNA used as probe
were labeled by random primer labeling technique with biotin-16-dUTP and
digoxigenin-11-dUTP respectively under supplier’s instructions. In situ
hybridization protocols as described by Fukui er al. (1994) were used with
minor modifications. Post- hybridization involved stringent washing to
the slides with 2 x SSC at 42°C and 0.1 x SSC at 60°C.
Somatic chromosome analysis showed that the two parents,
0. sativa and 0. officinalis as well as the F1 hybrid had 24 chromosomes.
Following FISH and using total genomic
|
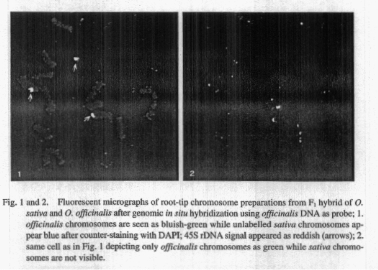
DNA of 0. officinalis as probe, parental chromosomes of officinalis could be distinguished in the F1 hybrid from the sativa chromosomes. The bluish-green fluorescein isothiocyanate (FITC) signal appeared on officinalis chromosomes when the hybrid cell was counter- stained by 4’-6-diamidino-2-phenylindole (DAPI) (Fig. 1). This is depicted more clearly when the same cell was exposed to blue light and only officinalis chromosomes were seen as green (Fig. 2). Hybridization with 45S rDNA probe, showed reddish fluorescent signal of rhodamine on two chromosomes of 0. officinalis and on one chromosome of 0. sativa (arrows in Fig. 1). Ohmido and Fukui (1997) also used multi-color FISH to identify simultaneously A genome-specific tandem repeat sequences and telomeric nucleotide sequences on rice prometaphase chromosomes. The results show that FISH can be conveniently used to differentiate parental chromosomes of sativa and officinalis in the F1 hybrid.
Fukui, K., N. Ohmido and G. S. Khush, 1994. Variability in rDNA loci in the genus Oryza detected through fluorescence in situ hybridization. Theor. Appl. Genet. 87: 893-899.