Recently an interest has been focused
on the nature and structure of the repetitive sequences surrounding functional
genes. Drastic changes in the repetitive sequences were demonstrated to
correlate with genome size among maize, sorghum and rice (Chen et a!. 1998).
However, the dynamic changes during the course of evolution have remained
largely unknown. The sequence analyses in non-coding regions suggest that
repetitive sequences tend to rapidly change during the course of evolution
in contrast to extensive preservation of both gene content and gene order
in the grasses. Although the importance of genome structure to gene regulation
is not clear, we intend to compare homologous regions including repetitive
sequences among related rice taxa. As the first step, the present study
was carried out to characterize molecular organization of the 260 kb region
in the vicinity of the waxy locus in rice. To grasp structural organization
focused on single copy regions around the waxy locus, the 260 kb restriction
fragments map constructed by using a bacterial artificial chromosome (BAC)
library from the Shimokita, a Japonica cultiver (Nagano eta!. 1997, Nagano
et a!. unpublished results) was used for this study.
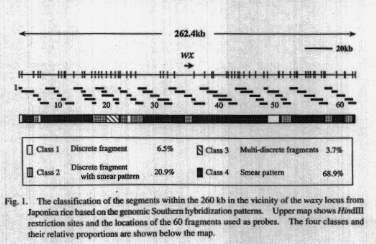
Overlapping 60 probes, which cover
the entire 260 kb region, were prepared from HindIII fragments and/or EcoRI
fragments. The probes ranging from 1.4 to 10 kb in fragment size were hybridized
with HindIll-digested Shimokita total DNA. The hybridization patterns obtained
were classified into four classes; the patterns indicating discrete band(s)
(class 1), multiple discrete bands without smear background (class 2),
discrete band(s) with smear background (class 3) and the patterns consisting
of solely smear bands (class 4). The characteristics of their patterns
and their distribution within the 260 kb are shown in Fig. 1. Each characteristic
is interpreted as follows.
Class 1: Four segments were classified into this class.
These sites showed a single or a very few bands in the Southern hybridization
patterns. A distribution of the four sites appeared to be spread in the
260kb region. The length of the four segments was 17 kb in total, corresponding
to 6.5% of the region. These segments should be solely present in the genome.
Class 2: About 55 kb in total length of ten segments
was regarded as the class 2. Interestingly, the 15 kb Hindill fragment
with the wx locus was observed as the class 2. Since this fragment contains
known Tnrl elements (Bureau et a!. 1996), the smear background in the pattern
of the class 2 might be caused by short repetitive elements. Alternatively,
a single copy region might be flanked by repetitive sequences. The positions
of the segment in this class, as well as the class 1, may contain genic
regions which have sequences for functional genes.
Class 3: In this class, one site gave rise to multiple
discrete bands without smear background. The patterns from the three probes
showed multiple copy fragments which are dispersed in a genome. This sort
of pattern was observed at a quite limited proportion (3.7%) of is genomic
region.
Class 4: The class 4 is a major characteristic around
the wx locus and possibly in other regions as well. About 70% of the 260
kb region showed to have strong smear background in the hybridization pattern.
The segments classified into this class are considered to have two possible
structures. One is a long repetitive sequence. Second is highly repetitive
short sequence(s) with an unique sequence.
References Bureau, T.E., P.C. Ronald and S.R. Wessler, 1996. A computer-based
systematic survey reveals the predominance of small inverted-repeat elements
in wild-type rice genes. Proc. Nati. Acad. Sci. USA 93:8524- 8529.
Chen, M. P. SanMiguel and J.L. Bennetzen, 1998. Sequence
organization and conservation in sh2/al-homolo- gous regions and rice.
Genetics 148: 435-443.
Nagano, H., 1. Wu, S. Kawasaki, Y.Kishima and Y. Sano, 1997.
Construction of a 300-kb BAC contig containing waxy locus in Japonica rice.
RON 14: 121-123.
|