J.Y. ZHUANG’, J.L. WU’, Y.Y. FAN’, H. LEUNG2 and K.L. ZHENG’
It has been demonstrated that resistance
gene analogs (RGAs) detected using denatured polyacrymide gel electrophoresis
can reveal high polymorphism in rice, offering an opportunity of using
RGA markers for a rapid germplasm identification in rice. However, the
feasibility of using RGA markers for germplasm studies would depend greatly
on whether the results revealed by RGA markers can represent the genetic
relatedness over the entire rice genome.
In our study, a common set of 16
indica rice varieties was used for RFLP and RGA analyses. To infer the
genetic relationships among the varieties, each polymorphic band was scored
as “1” for presence and “0” for absence. Genetic similarity between two
varieties was computed as the number of common bands divided by the total
number of bands in two varieties (Nei 1987, formula 5.53). The frequency
of polymorphism, computed as the number of polymorphic variety pairs divided
by the total number of variety pairs, was used to compare the polymorphism
detecting ability of polymorphic RGA and RFLP bands.
Ninety-seven RFLP probes were selected
to cover the 12 rice chromosomes based on published mapping information.
In combination with restriction endonuclease EcoR I, 39 of the 97 probes
detected polymorphism among the 16 varieties and produced a total of
90 polymorphic fragments. The average number of polymorphic
bands detected by each of the 97 probes was 0.93 only, and that over the
39 polymorphic probes was 2.3. All the varieties can be distinguished,
and the minimum number of polymorphic bands between any two varieties was
4. Over the 90 polymorphic bands, the average frequency of polymorphism
between all pairs of varieties was 0.299.
Four pairs of RGA primers were used
and generated 111 polymorphic bands among the 16 varieties. All the varieties
can be distinguished, and the minimum number of polymorphic bands between
any two varieties was 11. The average number of polymorphic bands detected
by one pair of RGA primer was 27.75. Over the 111 polymorphic bands, the
average frequency of polymorphism between all variety pairs was 0.376.
In addition, the number of polymorphic bands generated by different primer
pairs ranged as 28—34, and the average frequency of polymorphism ranged
as 0.365—0.395. This indicated that different primer pairs have similar
polymorphism detecting ability. Although the distribution of all RGA markers
was unknown and some RGA markers may be located as clusters, we could expect
that a small number of RGA primers would produce sufficient information
for germplasm evaluation in rice because of the great ability of RGAs on
detecting polymorphism.
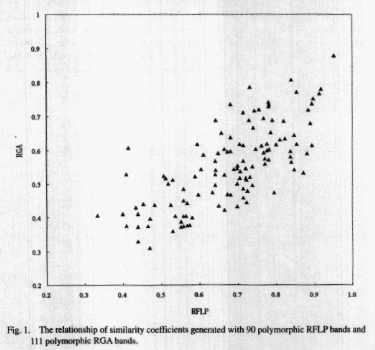
The value of similarities between
all pairs of varieties were computed based on the 90 polymorphic RFLP fragments
and the 111 RGA bands, respectively. The similarity coefficients generated
by RGA markers were plotted against those generated by RFLP markers (Fig.
1). Highly significant correlation (r=0.73) was obtained, indicating that
the cultivar relatedness revealed by four pairs of RGA primers was comparable
to that revealed by RFLP markers distributed over a large proportion of
rice genome. In addition, the genetic similarities revealed by RGA markers
were generally lower than that revealed by
RFLP markers (Fig. 1), indicating that RGA might be more
efficient for detecting polymorphism.
References Nei, M., 1987. Molecular evolutionary genetics. Columbia
University Press, New York, pplO6.
|