|
29. Identification of overlapping BAC clones carrying the
xa13 locus in rice
A.C. SANcHEZ1, L. ILAG2, D. YANG’, D.S. BRAR1, F. AUSUBEL2,
G.S. KHUSH’, M. YANO3,T. SASAKI3,
N. HUANG1,4 and Z. Li1
setts General Hospital, Boston, Massachusetts 02114, USA
We report the genetic and physical mapping of the recessive
gene xa13, which confers specific resistance to Philippine race 6 (PX099)
of Xoo. Resistance conferred by recessive genes like xa13 may represent
very different and largely unknown biochemical pathways in the host defense
system. Nine selected DNA markers and two F2 populations (NIL cross n =
131 and NPT cross n = 230) were used to construct a genetic map of the
xa13 region of rice chromosome 8. Four DNA markers, RG136, R2027, S14003
and Gi 149, closely flanking xa13 were used to identify bacterial artificial
chromosome (BAC) clones potentially harboring the xa13 locus from a rice
BAC library constructed using IR64 (Yang et al. 1997). All candidate BAC
clones identified by colony hybridization were subjected to Southern hybridization
analysis to confirm their overlaps. Clones were built into contigs based
on Hindffi fmgerprint analysis using digested clones as probes. Additional
probes for chromosome walking and contig orientation were generated by
TAILPCR of the positive BAC clones detected from colony hybridization (Liu
and Whittier 1995).
Clear and distinct reactions to the Xoo race 6 were observed
among the parental lines and F2 plants. In the NIL F2 population, plants
segregated into 106 susceptible (S) and 25 resistant (R) while in the NPT
F2 population, the segregation was 166S : MR. both agreeing with the expected
3:1 Mendelian ratio. All RFLP markers in the xa13 region also agreed with
the expected ratio of 1:2:1 at the 5% level. Figure 1 shows the two genetic
maps containing xa13 and all the selected DNA markers used in the two mapping
populations. The linear order of RG136, R2027, R2662, S 14003, and GI 149
in our maps was consistent with that determined by Harushima et al. (1998).
Our results clearly indicated that of these markers, RG136 and R2027 were
the closest ones flanking the xal3 locus. Table 1 shows the probes used
to screen the IR64 BAC library by colony hybridization and the corresponding
positive BAC clones. Overlaps were confirmed by hybridization of the positive
clones with the RFLP markers. Insert ends of the identified BAC clones
were amplified by TAIL-PCR and used as probes for hybridization with each
clone in the contigs. The resulting alignment of the overlapping clones
in these contigs was determined based on DNA hybridization patterns using
the digested clones and TAILPCR products as probes.
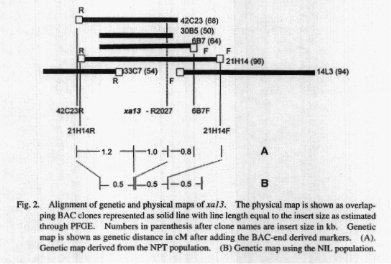
A single copy BAC end marker 42C23R, derived from the right
end of 42C23, was mapped 1.2 cM from xa13 while R2027 was 1 cM on the other
side using the NPT population. Another BAC-derived marker, 6B7F, is 0.8
cM beyond R2027. Two additional markers, 21H14F (left end) and 21H14R (right
end), derived from clone 21H14, were found to flank xa13 on either side
using the NIL population. These results indicated that
the contig and clone 21H14 contained the xa13 locus. The
genetic distance between the
insert ends of 2lHl4 was detennined to be 1 cM while the
physical size of 21H14 insert is
—96 kb, giving a physical/genetic ratio of 96 kb/cM in the
xa13 region of chromosome 8. Figure 2 shows the genetic map of xa13 aligned
with the physical map.
The construction of a contig map with overlapping BAC clones encompassing the xa13 locus represented a significant step toward our final goal to isolate this gene. Our results indicated that in the xa13 region of chromosome 8, each cM of genetic distance is roughly equivalent to 96 kb, a nearly 3-fold reduction in the ratio of physical/genetic map as compared to the average ratio of 260—280 kb/cM estimated by Wu and Tanksley (1993) and by Harushima et al. (1998). This reduction was apparently due to enhanced recombination in the region. The following steps to isolate xa13 including construction of the xa13 cDNA library, identification of the positive cDNA clones for the resistant allele of xal1, sub-cloning of clone 21H14, genetic complementation, etc., are underway. |

|