7. Identification of a null allele in traditional rice
germplasm of Taiwan, China
S.X. tang, G.S. kush and D.S. brar Division of Plant Breeding,
Genetics and Biochemistry IRRI, P.O. Box 933, 1099 Manila, Philippines
In isozyme analysis, those alleles that are not transcribed or that
code for detective polypeptides lacking enzymatic activity are referred
to as 'null alleles'. We analyzed 1604 genotypes of traditional rice germplasm
from Taiwan, China, through starch gel electrophoresis for 16 isozyme loci
following procedures of Glaszmann (1987). Null alleles were identified
for 5 isozyme loci, namely Cat-1, Adh-l, Est-1, Est-2, Est-5 at
a frequency of 0.25%, 0.12%, 4.93%, 54.61% and 0.19%, respectively. However,
a null allele for Est-9, which was not reported earlier, was identified
in 3 indica cultivars of Taiwan germplasm (Fig. 1).
54 Rice Genetics Newsletter Vol. 13
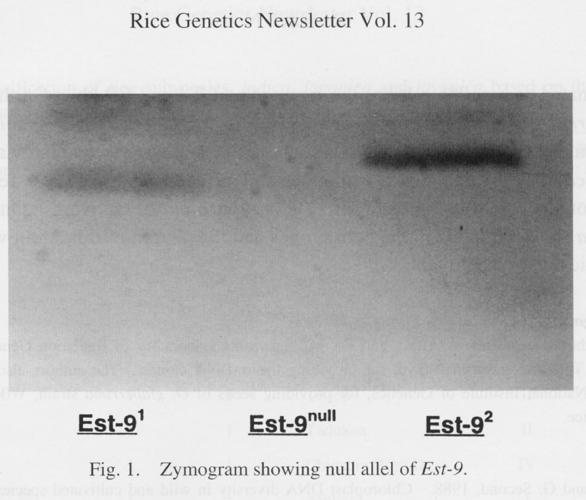 Est-9. located on chromosome 7 (Khush and Kinoshita
1991), has two alleles, Est-9^1 and Est-9^2 in proportion of 9.60% and
90.21%, respectively, in Taiwan rice germplasm. The null allele Est-9null
was identified in three cultivars, namely Ti-Chueh-Chu-Tzu (Acc. 65424),
Yun-Tzu-Ku (Ace. 65504) and Pao-Yeh-Chu (Ace. 78200). (Gene symbol: Old
system)
Est-9. located on chromosome 7 (Khush and Kinoshita
1991), has two alleles, Est-9^1 and Est-9^2 in proportion of 9.60% and
90.21%, respectively, in Taiwan rice germplasm. The null allele Est-9null
was identified in three cultivars, namely Ti-Chueh-Chu-Tzu (Acc. 65424),
Yun-Tzu-Ku (Ace. 65504) and Pao-Yeh-Chu (Ace. 78200). (Gene symbol: Old
system)
References
Glaszmann,J.C., 1987. Isozymes and classification of Asian rice varieties.
Theor Appl Genet 74: 31-20. Khush, G.S. and T. Kinoshita, 1991. Rice karyotype,
marker genes and linkage groups. Pages 83-1089, In
Rice Biotechnology, G.S. Khush and G.H. Toenniessen, eds., CAB
Intl., Oxford. U.K.
