III. Current linkage maps
Construction of current linkage maps are presented as shown in Fig.
1 and Table 3. In the linkage maps, the Kosambi mapping function was used
to correct the recombination values between genes. In the table, recombination
values are expressed as cM or percentages. Thus about 463 genes were allotted
to the twelve groups and 185 marker genes were positioned on twelve chromosomes.
Molecular linkage maps are already constructed in several groups
independently (Ref. 192, 219, 266, 385, 396, 539, 540. 572, 640, 644, 648,
650) and the integration of the different linkage maps has been progressed
(Ref. 91-95, 192, 193, 590, 620, 631, 651). Comparative mapping and construction
of synteny maps are progressed rapidly among different cereals and grasses
(Ref. 3, 5, 147, 220, 338, 575). Depending on the information of the position
of centromere, the orientation of the maps and allocation of genes to short
and long arms are going to be dissolved.
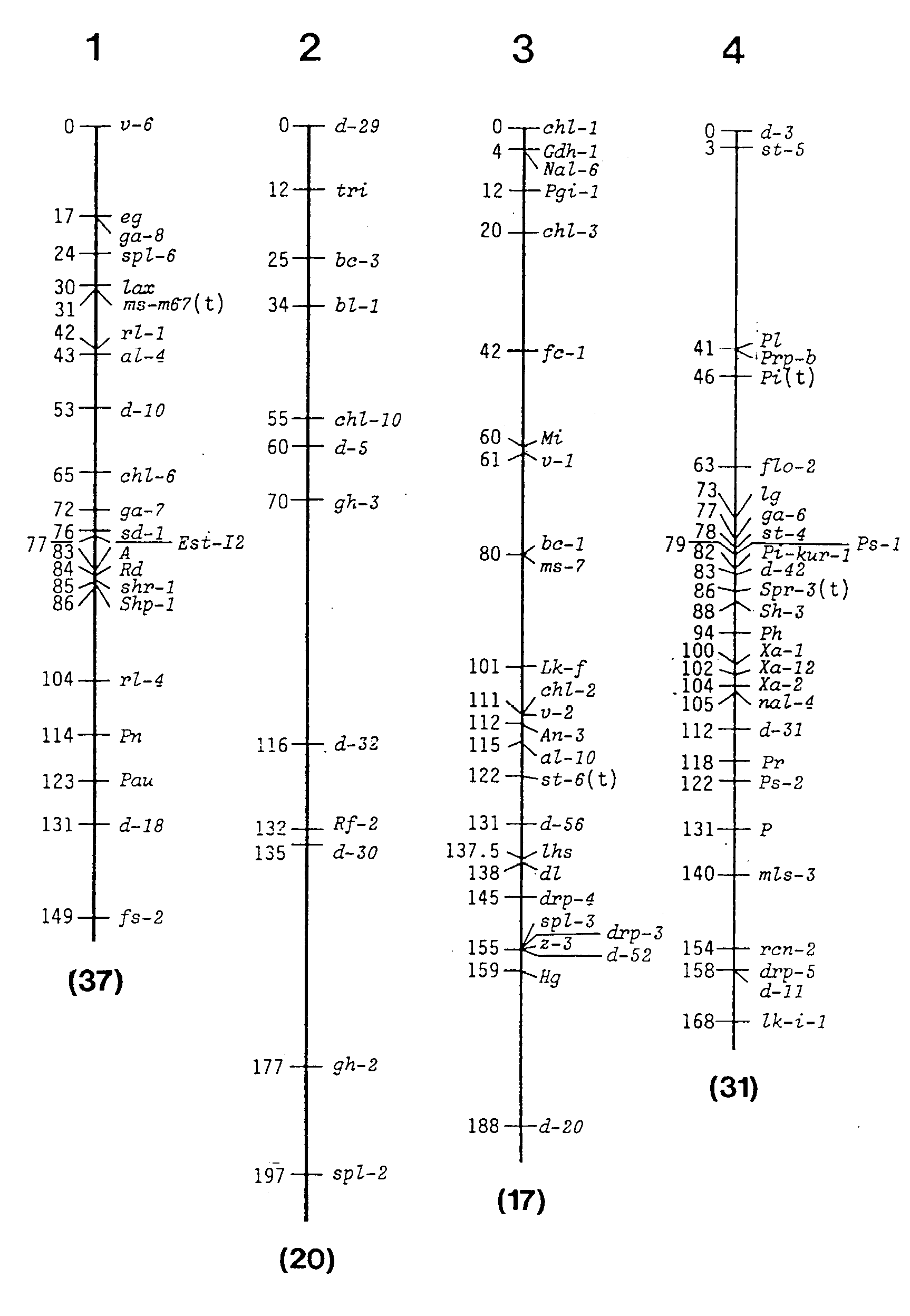


Fig. 1 1995 linkage maps of rice.
Figures in parentheses show the number of marker genes assigned to
the chromosome, but not given in the map.(as the exact location remains
undetermined).
Report of Committee on Gene Symbolization
Genes assigned to the linkage groups
| Chromosome 1 |
z-12 |
bc-2 |
S-A-1 |
z-4 |
Hbv |
| al-8 |
|
bd-I |
S-B-2 |
|
Lec |
| Ald-2 |
Chromosome 3 |
Cht-2 |
S-1 |
Chromosome 9 |
Pi-ta |
| Amy1B |
Actin-I |
Em |
S-8 |
Adh-2 |
Pi-6(t) |
| Arf-2 |
bl-4 |
er |
S-10 |
Amy3A/B/C |
Pro-1 |
| chl-5 |
Cat-AI |
Est-13 |
sbe-I |
chs-I(t) |
S-15 |
| d-2 |
d-14 |
eui |
Se-5 |
cyc-l |
|
| d-26 |
ga-2 |
gl-1 |
spl-4 |
d-57 |
|
| d-54 |
ga-3 |
Glh-6 |
Stv-a |
du-2120(t) |
|
| d-55 |
L |
glup-4 |
Un-a |
Est-3 |
|
| Dm-l |
Pms-2 |
1-Pl-1 |
Wc |
Est-12 |
|
| Est-5 |
Pox-cA |
M-Pox-1 |
Xa-7 |
Glup-l |
|
| Est-10 |
rl-5 |
Mdh-3 |
z-13 |
glup-2 |
|
| ga-9 |
s-e-I |
ms-14 |
zn |
gm-l |
|
| g.f-2 |
sh-4(t) |
0c-2 |
|
Man |
|
| Glu-l |
Shp-4 |
Pox-I |
Chromosome 7 |
ms-10 |
|
| Glu-1t |
st-3 |
Pro-2 |
Acp-4 |
R5s-3 |
|
| Glu-2t |
v-5 |
Pro-3 |
Beta-amy-1 |
R45s-l |
|
| Glu-3t |
v-7 |
Pro-5 |
d-7 |
|
|
| Glu-4t |
z-9 |
Rk-3 |
d-60 |
Chromosome 10 |
|
| glu-3(t) |
|
sd-7(t) |
ds-3(t) |
Bph-3 |
|
| Got-I |
Chromosome 4 |
sdg |
E-l |
bph-4 |
|
| lcd-l |
al-5 |
Shp-3 |
Est-I |
du-l |
|
| I-Ps-b |
a/-7(t) |
wgl(t) |
Est-7 |
ef(t) |
|
| lgt |
An-I |
xa-5 |
Est-9 |
Glh-3 |
|
| mp-l |
aul |
xa-13 |
ga-ll |
glu-2(t) |
|
| ms-m77(t) |
bph-2 |
ylb |
lp-i |
rDNA-2 |
|
| Oc-l |
drp-l |
z-7 |
Mal-I |
rk-2 |
|
| Prp-a |
drp-8 |
|
Mal-2 |
ygl |
|
| Rf-4 |
flo(t) |
Choromosome 6 |
ms-8 |
|
|
| S-13 |
ga-10(t) |
al-I |
Pms-I |
Chromosome 11 |
|
| S-16 |
ga-12 |
a/-9(t) |
Pox-3 |
D-2 |
|
| salT |
glup-3 |
Amp-5 |
Pox-4 |
esp-3 |
|
| sh-2 |
Gm-2 |
Amy2A |
Pro-4 |
Fdp-l |
|
| Tos-1 |
gpd-2 |
aph |
R5s-2 |
gal |
|
| tpi |
hpg-l |
bc-4 |
R.f-3 |
M-Pi-z |
|
| ts-a |
nal-I |
Cat-1 |
rl-6(t) |
nal-2 |
|
| z-8 |
nal-5 |
chl-7 |
S-7 |
PBR |
|
|
Pi-5(t) |
Cht-l |
S-9 |
Pi-f |
|
| Chromosome 2 |
Pin-1 |
Cht-3 |
se-2 |
Pi-is-1 |
|
| ae-3(t) |
pl-2W |
d-21 |
spl- 9 |
Pi-kur-2 |
|
| Amp-l |
rk-l |
drp-6 |
st-8 |
Pi-se-1 |
|
| AmyIA/C |
rl-2 |
du-2035(t) |
Un-b |
Pi-1 |
|
| bc-5 |
s-c-2 |
du-EM47(t) |
wp-l |
Pi-7(t) |
|
| cyc-2 |
s-e-2 |
dw-l |
Wph-l |
R5s-1 |
|
| D-l |
shp-5(t) |
Enp-l |
xa-k |
S-3 |
|
| Ef-x |
spr-l |
fc-2 |
z-10 |
Stv-b |
|
| Glu-5t |
ssk |
ga-4 |
|
Xa-3 |
|
| Glu-6t |
st-7 |
ga-5 |
Chromosome 8 |
Xa-4 |
|
| Glu-7t |
Wh |
Got-2 |
Amp-4 |
Xa-10 |
|
| Glu-8t |
Xa-14 |
1-PI-2 |
Amy3D/E |
Xa-a |
|
| Glu-9t |
ylm |
Ldh |
An-4(t) |
Xa-h |
|
| Glu-10t |
z-5 |
mp-2 |
chl-8 |
|
|
| glu-I(t) |
|
Pox-5 |
chl-9 |
Chromosome 12 |
|
| Got-3 |
Chromosome 5 |
PS |
d-51 |
Ald-3 |
|
| gpd-l |
Ald-1 |
rcn-5 |
shr-2 |
Bph-l |
|
| ms-17 |
An-2 |
s-a-I |
sk-2(t) |
Bph(t) |
|
| Pi-b |
Arf-l |
s-c-l |
ur-2 |
du-4 |
|
| z-11 |
ATP |
s-d-I |
xa(t) |
ga-13 |
|
Table 3. List of marker genes assigned to the respective linkage group
| Gene |
Name |
Gene locus |
Institution1 |
Literature No. |
| Group 1 |
|
|
|
|
| v-6 |
virescent-6 |
o |
KY |
126,139,146 |
| M |
extra glume |
17 5.3cM to XNpb97 |
KY |
129,137,146,620 |
| ga-8 |
gametophyte gene-8 |
17 |
HK |
304 |
| spl-6 |
spotted leaf-6 |
24 5.4cM to XNpbl74 |
KY |
136,137,146,620 |
| lax(lx) |
lax panicle |
30 6.6cM to XNpbll3 |
HK.KY |
129,137,146,292, 298,396,631 |
| ms-m67(t)* |
male sterile (Milyang
67ms) |
31 |
- |
491,492 |
| rl-1 |
rolled leaf-1 |
42 |
HK |
153,301 |
| al-4 |
albino-4 |
43 |
KY |
133,146 |
| d-10(d-15. d-16) |
kikeibanshinriki or
toyohikari-bunwaitillering dwarf |
53 |
KY.HK |
129,146,503 |
| chl-6 |
chlorina-6 |
65 |
KY |
126,139,146 |
| ga-7 |
gametophyte gene-7 |
72 |
HK |
242,249 |
| sd-l(d-47) |
dee-geo-woo-gen dwarf |
76 0.3cM to RG220 |
NA,HK |
10,29,30,49,179, 241,314,337,490,
565,566,631 |
| Est1-2* |
Esterase 1-2 |
77 OcM to RG220 |
- |
29,30 |
| A |
Anthocyanin activator |
83 4.4cM to XNpbl21 |
HK,KY |
146,192,281, 298,457 |
| Rd |
Red pericarp and seed
coat |
84 |
fs-2 |
181,298 |
| shr-1 |
shrunken endosperm-l |
85 |
KY |
262,442,602 |
| Shp-1 |
Sheathed panicle-1 |
86 |
HK |
153,244,256 |
| rl-4(rl-2) |
rolled leaf-4 |
104 9.4cM to XNpb368 |
HK.KY |
131,132,139,146, 262,620 |
| Pn |
Purple node |
114 |
HK.KY |
146,181,298 |
| Pau |
Purple auricle |
123 |
- |
30,274,480 |
| d-18(d-25)- |
hosetsu dwarf (d-18h)
of kotake-tamanishiki dwarf(d-18k) |
131 4.1cM to XNpb96 |
KY.HK |
126,134,139,146, 189,464,489,617,
620 |
| fs-2 |
fine stripe-2 |
149 8.7cM to XNpb216 |
HK.KY |
134,139,146,504, 620 |
| (Continued) |
|
|
|
|
| Gene |
Name |
Gene Locus |
Institution 1) |
Literature No. |
| Unlocated genes |
|
|
|
|
| al-8 |
albino-8 |
11%-d-18k |
KY |
133,139,146 |
| Ald-2* |
Aldolase-2 |
1.4cM-XNpbll3 |
NA |
50,196,396 |
| Amy1B* |
Alpha-amylase-1B |
1.0cM-RG146x |
IR |
365,376,386 |
| Arf-2* |
ADP-ribosylation factor (ARF)-2 |
2.1cM-0c-1 |
NA |
196 |
| chl-5 |
chlorina-5 |
13.2%-eg |
KY |
139,146 |
| d-2 |
ebisu dwarf |
15.6cM-XNpb359 |
HK,KY |
96,181,298 |
| d-26 |
7237 dwarf |
37%-A |
- |
81,83 |
| d-54(d-K-5) |
dwarf-Kyushu-5 |
25%-1ax |
KY |
139,146 |
| d-55(d-K-6) |
dwarf-Kyushu-6 |
13%-eg |
KY |
139,146 |
| Dm-l* |
Short culm showing dm type-1 |
39%-d-18k |
NG |
393 |
| Est-5 |
Esterase-5 |
triplo-1 |
IR |
369,589 |
| Est-10 |
Esterase-10 |
12.6CM-RG220 |
- |
18,19,20,640 |
| ga-9 |
gametophyte gene-9 |
l.3%-d-18k |
HK |
249 |
| gf-2 |
gold furrows of hull -2 |
triplo-1 |
IR |
401 |
| Glu-1(SrP) |
Rice glutelin-1 |
17%-sd-1 |
NA |
76,312,313,314, 350,508 |
| Glu-4t.lt.2t.3t* Glutelin
subunits (lt,2t,3t,4t) near XNpb343 |
Glutelin subunits (1t, 2t, 3t, 4t) |
near XNpb343 |
441 |
| glu-3(t)* |
lack of glutelin subunit-3 |
10.2cM-C250 |
NA |
99 |
| Got-1 |
Aspartate aminotransferase-1 |
triplo-1 |
IR |
369,589 |
| Icd-l |
Isocitrate dehydrogenase-1 |
triplo-1 |
IR |
369,589 |
| I-Ps-b |
Inhibitor for purple stigma |
linked with A |
- |
82 |
| lgt. |
long twisted grain |
16%-d-26 |
- |
81 |
| mp-l |
multiple pistil -1 |
triplo-1 |
IR |
228,367 |
| ms-m77(t)* |
male sterile(Milyang 77ms) |
15%-mp-l |
- |
491,492 |
| Oc-1* |
Oryzacystatin-1 |
l.lcM-XNpb92 |
NA |
196 |
| Prp-a(Pp) |
Purple pericarp |
7.2%-A |
HK |
84 |
| Rf-4*(Rf-2) |
Fertility restoration-4 |
near RG532 |
- |
235,545,637 |
| s-13 |
Hybrid sterility-13 (Pollen killer) |
near A |
Gl |
649 |
| s-16 |
Hybrid sterility-16 |
15 20%-Est-5 |
KT |
581 |
| salT* |
salt tolerance |
near CD0548 |
- |
35,540 |
| sh-2 |
shattering-2 |
l1%-sd-1 near
X Npbl74 |
NA |
314,323,337 |
| Tos-1* |
Retrotransposon-1 |
26%-sd-1 |
NA |
49,77 |
| tpi* |
triophosphate isomerase (cloned gene) |
near C146 |
NA |
50 |
| ts-a |
twisted stem |
23%-A |
- |
81 |
| (Continued) |
|
|
|
| Gene |
Name |
Gene Locus |
Institution 1) |
Literature No. |
| z-8 |
zebra-8 |
triplo-1 |
IR |
400,401 |
| Group 2 |
|
|
|
|
| d-29(d-K-l) |
short uppernost internode
dwarf |
0 |
KY.HK |
132,134 |
| tri |
triangular hull |
12 16.7cM to XNpb250 |
KY,HK |
130,146,296,620 |
| bc-3 |
brittle culm-3 |
25 |
KY |
132,134,145 |
| bl-1 |
brown leaf spot-1 |
34 10.3cM to XNpb250 |
HK.KY |
145,298,620 |
| chl-10 |
chlorina-10 |
55 |
KY |
145 |
| d-5 |
bunketsu-waito tillering
dwarf |
60 |
HK |
181,298 |
| gh-3 |
gold hull and internode-3 |
70 |
KY |
145 |
| d-32 (d-12. d-K-4) |
dwarf Kyushu-4 |
116 |
KY |
128,145 |
| Rf-2 |
Pollen fertility restoration-2 |
132 |
RY |
468,470,587 |
| d-30(d-W) |
waisei-shirasasa dwarf |
135 near XNpb227 |
KY.HK |
91,130,134, 145,631 |
| gh-2 |
gold hull and internode-2 |
177 |
KY.HK |
130,134,138,145, 401 |
| spl-2(bl-3) |
spotted leaf-2 |
197 9.2cM to XNpb349 |
KY |
92,134,137,145 |
| Unlocated genes |
|
|
|
|
| ae-3(t)* |
amylose extender-3 |
triplo-2 |
IR |
166 |
| Amp-l |
Aminopeptidase-1 |
triplo-2 19.4cM-XNpb223 |
IR.NA |
306,369,396,569 |
| AmylA/C* |
Alpha-amylase-IA/C |
9.2CN-RG95 |
IR |
365,376,386 |
| bc-5 |
brittle node |
triplo-2 |
IR |
173,401 |
| cyc2* |
cyclophilin-2 |
near RZ957 |
- |
540 |
| D-1* |
Dominant lethal from
Q.longistaminata |
39%-Amp-l |
IR |
57 |
| Ef-x* |
Earliness-x |
18%-TR2-9 |
RY |
426,434 |
| Glu-5t* |
Glutelin subunit-5t |
10%-XNpb227 |
KY |
441,574 |
| Glu-6t.7t.8t* |
Glutelin subunits
(6t,7t,8t) |
0.7%-Glu-5t 2.2%-XNpb21 |
KY |
441,574 |
| Glu-9t* |
Glutelin subunit-9t |
near XNpb227 |
KY |
441 |
| Glu-10t* |
Glutelin subunit-10t |
near Glu-9t |
KY |
441 |
| glu-1(t)* |
lack of glutelin subunit-1 |
near XNpb243 |
NA |
98,99 |
| Got-3 |
Asparate aminotransferase-3 |
triplo-2 |
IR |
369,453 |
| gpd-1* |
glyceraldehydephosphate
dehydrogenase-1 |
near C1061 |
NA |
50 |
| (Continued) |
|
|
|
Literature No. |
| Gene |
Name |
Gene locus |
Institution 1) |
| ms-17 |
male sterile-17 |
35%-gh-2 |
KT |
206,207 |
| Pi-b(Pi-s) |
Pyricularia oryzae
resistance-b |
5.8%-TR2-10 OcM-R1792 |
NA |
71,203,277, 451,471 |
| z-11 |
zebra-ll |
trlplo-2 |
IR |
400,401 |
| z-12 |
zebra-12 |
triplo-2 |
IR |
400,401 |
| Group 3 |
|
|
|
|
| chl-l |
chlorina-1 |
0 7.1cM to XNpbl64 |
KY |
130,134,144,358, 396,620 |
| Gdh-l |
Glutametedehydrogenase-1 |
4 |
Gl |
115,120,122, 123,287 |
| Nal-6 |
Narrow leaf-6 |
4 |
KY |
144 |
| Pgi-1 (Pgi-a) |
Phosphoglucose isomerase-1 |
12 near C217 |
IR,GI |
120,122,123, 322,374.375 |
| chl-3 |
chlorina-3 |
20 |
KY |
144,358 |
| fc-1 |
fine culm-l |
42 10.9cM to XNpb249 |
KY |
132,134,138,144, 358,620 |
| Mi |
Minute grain |
60 |
HK |
512,517,524, 527,528 |
| v-1 |
virescent-1 |
61 |
KY,HK |
132,134,144,148, 149,358 |
| bc-l |
brittle culm-1 |
80 |
HK.KY |
144,298 |
| ms-7 |
male sterile-7 |
80 |
KT |
206,207 |
| Lk-f |
'Fusayoshi' long grain |
101 |
HK, OK |
256,512,516. 517,527,529 |
| chl-2 |
chlorina-2 |
III |
KY |
134,136,358 |
| v-2. |
virescent-2 |
III |
KY |
134,140,144,358 |
| An-3 |
Awn-3 |
112 |
HK |
298,421,517,524 |
| al-10 |
albino-10 |
115 |
KY |
140,144 |
| st-6(t) |
stripe-6 |
122 |
HK |
254 |
| d-56(d-K-7) |
dwarf Kyushu-7 |
131 |
KY |
140,144 |
| lhs (lhs-2,op) |
leafy hull sterile |
137.5 |
KY,HK,IR |
80,127,170,191, 256 |
| dl(lop) |
drooping leaf |
138 16.6cM to XNpb394 |
KY,HK |
130,137,138,144, 192,256,282,358 |
| drp-4 |
dripping-wet leaf-4 |
145 |
KY |
140,141,144 |
| scl-3(bl-14) |
spotted leaf-3 |
155 15.OcM to XNpbl73 |
KY |
134,137,138,144, 620 |
| drp-3 |
dripping-wet leaf-3 |
dl(lop) |
KY |
140,141,144 |
| (Continued) |
|
|
|
|
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| z-3 |
zebra-3 |
155 |
KY |
136,140,144 |
| d-52(d-K-2) |
dwarf Kyushu-2 |
155 |
KY |
136,140,144 |
| Hg |
Hairy glume |
159 near RG348 |
HK |
182,298,299,631 |
| d-20 |
hayayuki dwarf |
188 |
HK |
174,189,517,524 |
| Unlocated genes |
|
|
|
|
| Actin-1* |
Actin-1 |
near CD0337 |
- |
540 |
| bl-4 |
brown leaf spot-4 |
29%-bc-l |
HK |
504 |
| Cat-AI* |
Catalase-AI (cDNA
clone) |
1.4cM-XNpb279 |
NA |
195 |
| d-14(d-10) |
kanikawa-bunwai tlllering
dwarf |
32%-dl |
HK |
503 |
| ga-2 |
ganetophyte gene-2 |
11%-dl |
NA.KY |
302,308 |
| ga-3 |
gametophyte gene-3 |
34%-dl |
NA.KY |
302,308 |
| L* |
Lethal factor |
0.2gcM-XNpb23 |
NA |
28 |
| Pms-2* |
Photoperiod-sensitive
male sterility-2 |
near RG191 |
- |
635 |
| Pox-cA* |
Peroxidase-cA (cloned
gene) |
l.lcM-XNpb244-2 |
NA |
194 |
| rl-5(rl-3) |
rolled leaf-5 |
13%-chl-l |
KY |
140 |
| s-e-1 |
hybrid sterility-e |
16%-bc-l |
Gl |
346 |
| sh-4(t)* |
shattering-4 |
near XNpb40 |
NA |
52 |
| Shp-4(Gb) |
Sheathed panicle-3 |
27%-bc-l |
HK |
73,474 |
| st-3(stl) |
stripe-3 |
1.1%-bc-l |
KY |
140 |
| v-5 |
virescent-5 |
2.0%-chl-l |
KY |
141,144,358 |
| v-7 |
virescent-7 |
1. 7%-bc-l |
KY |
140 |
| z-9 |
zebra-9 |
13cM-Pgi-l |
IR |
382,400,401 |
| Group 4 |
|
|
|
|
| d-3 |
bunketsu-waito tillering
dwarf |
0 |
HK |
181,298 |
| st-5 |
stripe-5 |
3 |
HK |
246,256 |
| Pl |
Purple leaf |
41 |
HK.KY |
185,298,300,501 |
| Prp-b(Fb) |
Purple pericarp |
41 19.0cM to RG329 |
- |
84,642 |
| Pi(t) |
Pyricularia oryzae
resistance |
46 |
- |
83 |
| flo-2 |
floury endosperm-2 |
63 |
HK.KY |
166,243,443 |
| lg |
1iguleless |
73 3.4cM to XNpbl97 |
HK.KY |
247,256,298, 396,620 |
| ga-6 |
ganetophyte gene-6 |
77 |
HK |
242,252 |
| st-4(ws-2) |
stripe-4 |
78 |
HK |
242 |
| Gene |
Name |
Gene Locus |
Institution 1) |
Literature No. |
| Ps-1 |
Purple stigma-1 |
79 |
Gl |
348,500 |
| {�i-Kur-I* |
Pyricularia oryzae
resistance-kur-1 |
82 |
YA |
61 |
| d-42 |
liguleless dwarf |
83 |
HK |
85,183 |
| Spr-3(t) |
Spreading panicle-3 |
86 |
Gl |
39,414 |
| Sh-3 |
Shattering-3 |
88 |
Gl |
39,414 |
| Ph=Bh-c(Po) |
Phenol staining |
94 0.lcM to V17 |
HK,KY,GI |
7,221,231,297, 377,396,540,640 |
| Xa-l(Xe) |
Xanthomonas campestris
pv. oryzae resistance-1 |
100 0cM to Y03700 |
NA |
397,594,623, 624,626.627 |
| Xa-12(Xar-kg) |
oryzae resistance-12 |
102 |
NA |
326,593,594 |
| Xa-2 |
Xanthomonas campestris
pv. oryzae resistance-2 |
104 2.0cM to XNpb264 |
NA |
43,397,626 |
| nal-4(nal) |
narrow leaf-4 |
105 |
|
611 |
| d-31 |
tai chung-155 irradiated
dwarf |
112 |
- |
611 |
| Pr |
Purple hull |
118 16.8cM to RG163 |
HK.KY |
181,298,631 642 |
| Ps-2 |
Purple stigma-2 |
122 |
Gl |
81,82,348 |
| P |
Colored apiculus |
131 |
HK |
281,298,348,457 |
| 1118-3 |
malformed spikelet-3 |
140 |
HK |
511,518,522 |
| rcn-2 |
reduced culm number-2 |
154 |
HK |
256,511,522,523 |
| drp-5 |
dripping-wet leaf-5 |
158 |
KY |
445,447 |
| d-ll (d-g) |
shinkane-aikoku or
nohrin-28 dwarf |
158 11.0cM to XNpb49 |
KY.HK |
130,298,620,631 |
| lk-i -l |
IRAT 13' long grain |
168 |
HK |
187,256,515, 522,530 |
| Unlocated genes |
|
|
|
|
| al-5 |
albino-5 |
34%-lg |
KY |
133 |
| al-7(t) |
albino-7 |
31%-lg |
KY |
133 |
| An-1 |
Awn-1 |
5. 4%-d-ll |
HK |
298 |
| aul |
auricleless |
triplo-4 |
IR |
228 |
| bph-2 |
brown planthopper
resistance-2 |
near Bph-l (chr.l2) |
NA,IR |
100,101,476 |
| drp-l |
dripping-wet leaf-1 |
39%-d-2(chr.l) |
HK |
301 |
| drp-g |
dripping-wet leaf-8 |
28%-lg |
KY |
447 |
| flo(t)* |
floury endosperm |
triplo-4 |
IR |
166 |
| ga-10(t) |
gametophytegene-10 |
27%-lg |
HK |
ga-10(t) |
| (Continued) |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| ga-12 |
gametophyte gene-12 |
2.1%-shp-5(t) |
|
|
| glup-3 (esp-7) |
glutelin precursor-3 |
trisomic-4 |
KY |
125,449 |
| Gm-2 |
Gall midge resistance-2 |
1.3 cM-RG329 |
IR |
26,178,279,395 |
| gpd-2* |
glyceraldehydephosphate
dehydrogenase-2 |
near C377 |
NA |
50 |
| hpg-l* |
high preproglutelin-1 |
13.3cM-L102 |
NA |
98 |
| nal-l |
narrow leaf-1 |
25%-d-2(chr.1) |
HK |
282 |
| nal-5(nal-l) |
narrow leaf-5 |
9.5%-lg |
KY |
132,134 |
| Pi-5(t) |
Pyricularia oryzae
resistance-5 |
near RG788 |
IH |
582 |
| Pin-1 |
Purple internode-1 |
31%-Pl |
HK |
284 |
| pl-2(t) |
purple leaf-2 |
triplo-4 |
IR |
401 |
| rk-1 |
round kernel-1 |
35% -lg |
KY.HK |
131,132,134 |
| rl-2 |
rolled leaf-2 |
35%-d-2(chr.1) |
HK |
282 |
| s-c-2 |
hybrid sterility-c |
31%-Ph |
Gl |
344 |
| s-e-2 |
hybrid sterility-e |
15%-lg |
Gl |
346 |
| shp-5(t) |
sheathed panicle-5 |
near d-2(chr.1) |
HK |
250 |
| spr-1 |
spreading panicle-1 |
27%-Pl |
HK |
186 |
| ssk(sk) |
malformed semisterile |
6.8%-Pl |
- |
66 |
| st-7 |
stripe-7 |
triplo-4 |
IR |
401 |
| Wh |
White hull |
8.0%-lg |
HK |
150,298 |
| Xa-14 |
Xanthomonas campestris
pv oryzae resistance-14 |
|
NA |
541 |
| ylm |
yellow leaf margin |
10%-lg |
KY |
132,134 |
| z-5 |
zebra-5 |
11%-lg |
HK |
184 |
| Group 5 |
|
|
|
|
| gh-1 |
gold hull and internode-1 |
0 |
HK.KY |
144,298 |
| nl-2 |
neck leaf-2 |
6 |
KY |
132,134,143 |
| d-l |
daikoku dwarf |
30 near XNpb105 |
HK.KY |
95,143,181,276, 298,617 |
| st-2(gw) |
stripe-2 |
49 |
HK |
298 |
| al-3 |
albino-3 |
51 |
KY |
133,143 |
| spl-8(bl-8) |
spotted leaf-8 |
51 |
KY |
134,137,143 |
| al-6(t) |
albino-6 |
56 |
KY |
133 |
| flo-l |
floury endosperm-l |
63 |
KY |
166,443,444 |
| ops |
open hull sterile |
69 |
KY |
129,134,143 |
| v-10 |
virescent-10 |
77 |
KY |
444,446 |
| (Continued) |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| bg1 |
bright green leaf |
82 |
KY |
134,141,143 |
| ri |
verticillate rachis |
85 |
HK.KY |
129,143,298,617 |
| spl-7 |
spotted leaf-7 |
100 near XNpb81 |
KY |
95,134,136,137, 143 |
| nl-1 |
neck leaf-1 |
110 0cM to XNpb188 |
HK.KY |
143,192,298, 431,621 |
| al-2 |
albino-2 |
111 |
KY |
133,137,143 |
| Unlocated genes |
|
|
|
|
| Ald-1* |
Aldolase-1 |
0.8cM-Cht-2 7.7cM-C13 |
NA |
50,196,396 |
| An-2 |
Awn-2 |
33%-gl-l |
HK |
298,422 |
| Arf-1* |
ADP-ribosylation factor
(ARF)-1 |
1. 7cM-Oc-2 |
NA |
196 |
| ATP* |
ATPase |
near RG697 |
- |
540 |
| bc-2 |
brittle culm-2 |
triplo-5 |
IR |
401,504 |
| bd-1 |
beaked lemma |
22%-gI-1 |
- |
274,422 |
| Cht-2* |
Chitinase-2 |
2.9cM-XNpbl05 |
NA |
320,396 |
| Em* |
ABA-regulated gene/expressed
in embryogenesis |
trisomic-5 |
IR |
376 |
| er(o) |
erect growth habit |
38%-gh- 1 |
HK |
504 |
| Est-13 |
Esterase-13 |
17.9cM-RG13 |
- |
19 |
| eui |
elongated uppermost
internode |
27%-nl-l |
HK |
248,253,390, 391,643 |
| gl-1 |
glabrous leaf and
hull |
12%-TRI-5d 10.5cM-XNpb396 |
HK.KY |
298,431,621,631 |
| Glh-6 |
Green leafhopperresistance-6 |
tripro-5 |
IR |
383,550 |
| glup-4 |
glutelin precursor-4 |
trisomic-4 |
KY |
125 |
| l-Pl-1 |
Inhibitor for purple
leaf-1 |
31%-gh-1 |
HK |
181,298,300 |
| M-Pox-1 |
Modifier for Pox-1 |
near gl-l |
CH |
360,362 |
| Mdh-3* |
Malate dehydrogenase-3 |
MAAL 5 |
IR |
177 |
| ms-14 |
male sterile-l4 |
11%-nl-1 |
KT |
205,206 |
| Oc-2* |
Oryzacystatin-2 |
0.4cM-XNpb255 |
NA |
196 |
| Pox-1 |
Peroxidase-1 |
38%-nl-1 |
CH.GI |
288,362,363 |
| Pro-2* |
Prolamin-2 |
6.8%-XNpb251 |
KY |
441 |
| Pro-3* |
Prolamin-3 |
2.2%-Pro-2 |
KY |
441 |
| Pro-5* |
Prolamin-5 |
3.0%-Pro-3 |
KY |
441 |
| Rk-3 |
Round Kernel-3 |
triplo-5 |
IR |
401 |
| sd-7(t) |
semidwarf-7 |
O%-d-1 |
CH |
562, 564 |
| Gene |
Name |
Gene locus |
Institution |
Literature No. |
| sdg* |
semidwarf (BRGPC) |
4.3cM-RZ182 |
|
227,638 |
| Shp-3(Ga) |
Sheathed panlcle-3 |
37%-nl-l |
- |
73,474 |
| wgl(t)* |
spikelet width |
OCM-W168A |
NA |
157 |
| xa-5 |
Xanthomonas campestris
pv oryzae resistance-5 |
trisomic-5 near gl-l.
and XNpb396 |
IR RG207 |
267,368,478, 484,618,621 |
| xa-13 |
Xanthomonas campestris
pv oryzae resistance-13 |
9. 7%-xa-5 |
IR |
331,394 |
| ylb |
yellow banded leaf
blade |
32%-nl-l |
- |
82 |
| z-7 |
zebra-7 |
triplo-5 |
IR |
400,401 |
| Group 6 |
|
|
|
|
| d-4 |
bunketsu-waito tillering
dwarf |
0 |
HK |
181,298 |
| I-Pl-4 |
Inhibitor for purple
pericarp |
9 |
HK |
181,187 |
| wx(am) |
glutinous endosperm |
22 l.lcM to C425A |
HK.KY |
74,91,93,225, 298,405,417,
443,540 |
| dp-l |
depressed palea-1 |
24 10.8cM to XNpb209 |
HK.KY |
94,130,294 |
| ms-1 |
male sterile-1 |
27 |
- |
66 |
| En-Se-1 |
Enhancer for photosensitivity
(Se-I) |
28 |
Gl |
410,412 |
| Pgd-2 |
Phosphogluconate dehydrogenase-2 |
29 |
GI,IR |
117,122,123, 177,287,453 |
| v-3 |
virescent-3 |
30 |
KY |
134,358 |
| ga-1 |
gametophyte gene-1 |
36 |
KY |
135,309 |
| lcr |
Low crossability |
41 |
Gl |
411,412 |
| C |
Chromogen for anthocyanin |
44 16.4cM to XNpbl65-l |
HK |
181,192,298, 317,457 |
| S-5 |
Hybrid sterility-5 |
48 4.4cM to RG213 |
NA.KT |
103,104,105, 579,598 |
| Amp-3(Amp-l) |
Aminopeptidase-3 |
53 1.7%-RG213 |
GI.IR |
16,122,123,369, 403 |
| Est-2 |
Esterase-2 |
54 1.7% to RG213 |
NA.IR |
16,18,303,305, 307.369,579,698 |
| bl-3 |
brown leaf spot-3 |
55 |
HK |
604 |
| alK |
alkali degeneration |
56 1.1 cM to C1478 |
KY.NA |
210,231,598 |
| st-l(ws) |
stripe-1 |
65 |
KY.HK |
294 |
| Gene |
Name |
Gene locus |
Institution |
Literature No. |
| Pgi-2 (Pgi-b) |
Phosphoglucose isomerase-2 |
67 7.9% to RG213 |
GI,IR |
16,122,123,322, 359,369,398,
403,598 |
| Se-l (Lf, Lm, Rs) |
Photosensitivity-1 |
67 4.7%. to XNpb294
and 0.3cM to C235 |
NA |
22,48,210,339, 340,341,355,359,
534,540,598,613, 614,615 |
| Pi-z (Pi-2) |
Pyricularia oryzae
resistance-z |
69 2.9cM to XNpb294
and 2.1cM to RG64 |
NA |
48,64,78,108, 110,201,203,
237.271,451, 452,471,630 |
| S-6 |
Hybrid sterility-6 |
74 |
Cl |
407,409 |
| d-9 |
Chinese dwarf |
76 |
HK |
181,187 |
| rcn-1 |
reduced culm number-1 |
77 |
HK |
513,522 |
| gf-1 |
gold furrows of hull-1 |
80 |
HK |
152 |
| chl-4 |
chlorina-4 |
82 |
KY |
134,358 |
| bl-2(bl-m) |
brown leaf spot-2 |
83 |
HK |
150,301 |
| fs-1 |
fine stripe-1 |
95 |
HK |
190,298 |
| Cl |
Clustered spikelets |
96 |
HK |
150,298,367 |
| ms-9 |
male sterile-9 |
100 |
KT |
206,425,429 |
| HI-a |
Hairly leaf |
118 |
HK |
182,298,299.522 |
| Ur-1 |
Undulate rachis-1 |
125 |
HK |
181,298 |
| d-58 |
small grained dwarf |
139 |
HK |
514,517,522 |
| Unlocated genes |
|
|
|
|
| al-l |
albino-1 |
7.1%-wx |
KY |
133 |
| al-9(t) |
albino-9 |
trisomic-6 |
KY |
141 |
| Amp-5 |
Aninopeptidase-5 |
15.OcM-Est-2 |
- |
17 |
| Amy2A* |
Alpha-amylase-2A |
6.3cM-HG433 |
- |
365,376,386 |
| aph |
apiculus hairs |
linked with Est-2
and Pgi-2 |
GI |
436,438 |
| bc-4 |
brittle culm-4 |
triplo-6 |
IR |
228 |
| Cat-1 |
Catalase-1 |
22%-Pox-5 8.1cM-BG244 |
IR,GI |
118,120,589,640 |
| chl-7 |
chlorina-7 |
27%-Pi-z 13%-Pgi-2 |
NA |
228,357,399 |
| Cht-I* |
Chitinase-1 |
0.4cM-XNpb342 |
NA |
320,396 |
| Cht-3* |
Chitinase-3 |
0.4cM-XNpb342 |
NA |
320,396 |
| d-21 |
aomorimochi-14 dwarf |
8.3%-wx |
HK |
189 |
| drp-6 |
dripping-wet leaf-6 |
15%-fc-2 |
KY |
445,447 |
| du-2035* |
dull endosperm
(2035) |
triplo-6 |
IR |
15%-fc-2 |
| (Continued) |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| du-EM47* |
dull endospern (EM47) |
tri plo-6 |
IR |
166 |
| dw-l(fh) |
deep water tolerance-1 |
30%-Se-l |
- |
149,373 |
| Enp-l* |
Endopeptidase-1 |
2.2%-Cat-l |
IR |
369 |
| fc-2 |
fine culm-2 |
18%-C |
KY |
445,447 |
| ga-4(ga-A) |
gametophyte gene-4 |
34%-wx |
HK.KY |
283,309,310 |
| ga-5(ga-B) |
gametophytegene-5 |
27%-wx |
HK |
283 |
| Got-2 |
Aspartateam inotranspherase-2 |
31%- Pgi-2 |
GI,IR |
369,453 |
| 1-Pl-2 |
Inhibitor for purple
leaf-2 |
10%-1-Pl-4 |
HK |
181,187,300 |
| Ldh* |
Lactate dehydrogenase |
0.9cM-C1003B |
NA |
22B.226 |
| mp-2 |
multiple pistil-2 |
triplo-6 |
IR |
228.366 |
| Pox-5 |
Peroxidase-5 |
39%-Pgi-2 |
IR |
589 |
| PS* |
Photoperiod sensitivity |
23%-Pgi-2 0cM-RG64 |
IR |
239,398 |
| rcn-5 |
reduced culm number-5 |
22%-C |
HK |
520 |
| s-a-1 (s1,
x1 |
hybrid sterility-a |
21%-wx |
Gl |
343,346 |
| s-c-l |
hybrid sterility-c |
8.6%-C |
Gl |
344 |
| s-d-l |
hybrid sterility-d |
33%-wx |
GI |
346 |
| S-A-I(A-I) |
Hybrid sterillty-A |
9.5%-C |
Gl |
347,349 |
| S-B-2(B-2) |
Hybrid sterility-B |
28%-wx |
Gl |
347,349. |
| S-1 |
Hybrid sterility-1 |
near C |
Gl |
408,416 |
| S-8 |
Hybrid sterility-8 |
ll%-Cat-1 |
KT |
579,596,597 |
| S-10 |
Hybrid sterility-10/gamete
eliminator |
0.16%-wx |
Gl |
418,420 |
| sbe-1* |
starch branching enzyme-1 |
0.3cM-G342 |
NA |
168,316 |
| Se-5 |
Photosensitivity-5 |
27%-Pi-zt 23cM-Pgi-2 |
NA |
616 |
| spl-4(bl-15) |
spotted leaf-4 |
2.5%-dp-l 5.0cM-XNpb209 |
KY |
94,134,137 |
| Stv-a(St) |
Stripe disease resistance-a |
38%-wx |
NA |
584,585,586 |
| Un-a |
Uneven grain |
22%-Cl-a |
- |
548 |
| Wc* |
Wide compatibility |
5.9cM-Est-2 |
IR |
216,234 |
| Xa-7 |
Xanthomonas campestris
pv oryzae resistance-7 |
8.8X-Q1091 |
NA |
156,324,478,484 |
| z-13 |
zebra-13 |
triplo-6 |
IR |
400,401 |
| zn |
zebra necrosis |
20%-C |
HK |
184,256 |
| Group 7 |
|
|
|
|
| d-6 |
ebisumochi or tankanshirasasa
dwarf |
0 near XNpb50 |
HK.KY |
0 near XNpb50 |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| g-1 |
long sterile lemmas-1 |
6 24.9cM to XNpb85-l |
HK.KY |
92,95,141,298, 518 |
| spl-5(bl-6) |
spotted leaf-5 |
31 near XNpb85-l |
KY |
95,132,137,141 |
| Rc |
Brown pericarp and
seed coat |
44 OcM to C67A |
HK.KY |
141,181,231,298, 505,631 |
| v-11 |
virescent-11 |
45 near XNpb85-l |
KY |
95,444,446 |
| z-6 |
zebra-6 |
47 |
HK |
245,256 |
| rfs |
rolled fine striped
leaf |
58 1.4cM to XNpb20 |
HK.KY |
134,141,192,256 |
| slg |
slender glume/mutator |
64 3.0% to XNpb33 |
KT |
209,546 |
| ge |
giant embryo |
79 |
KY.HK |
79,134,442,444 |
| esp-l(rsp-1) |
endosperm storage
protein-1 |
88 |
KY |
211,212,213,215, 440 |
| Unlocated genes |
|
|
|
|
| Acp-4 |
Acid phosphatase-4 |
near R2387 |
IR |
65,540 |
| Beta-Amy-1 |
Beta-amylaseisozyme-1 |
trsomic-7 |
KY |
472,473 |
| d-7 |
heiei-daikoku or cleistogamous
dwarf |
39%-d-6 |
HK |
181,298 |
| d-60 |
dwarf (Hokuriku 100) |
trisomic-7 |
KT |
537,553 |
| ds-3(t) |
desynapsis-3 |
tripro-7 |
KY |
197,609 |
| E-1=m-Ef-1* |
Earliness-1 |
trisomic-7
23%-Rc |
KT,CH |
351,352,354,355 498,558,559 |
| Est-1 |
Esterase-1 |
triplo-7 |
NA,IH |
16,65,303,306, 369 |
| Est-7 |
Esterase-7 |
24%-Acp-4 |
IR |
65,453 |
| Est-9 (Est-cl) |
Esterase-9 |
triplo-7 0.4%-Rc |
GI,IR |
229,369,374, 375,403 |
| ga-ll |
gametophytegene-ll |
23%-Est-9 |
KT |
229,230 |
| lp-1 |
long palea-1 |
12%-Un-b |
- |
548 |
| Mal-l |
Malate dehydrogenase
(MADP)-1 |
20.3cM-RGS28 |
gi.ir |
16,114,540,640 |
| Mal-2 |
Malate dehydrogenase
(MADP)-2 |
1.8cM-Ra511 |
- |
21,640 |
| ms-8 |
male sterile-8 |
20%-rfs |
KT |
205,206,207 |
| Pms-1* |
Photoperiod-sensitive
malesterility-1 |
4.3cM-RG477 |
- |
635 |
| Pox-3 |
Peroxidase-3 |
5%-Pox-4 |
IR |
361,369 |
| Pox-4 |
Peroxidase-4 |
31%-Est -l |
IR |
369,453 |
| Pro-4* |
Prolamin-4 |
1.6%-XNpb338 |
KY |
441 |
| Gene |
Name |
Gene locus |
Institution 1) |
Liiterature No. |
| R5s-2* |
5s ribosomal DNA-2 |
short arm |
NA |
33 |
| Rf-3*(Rf-l) |
Fertility restoration-3 |
near XNpb379 |
- |
235,545,637 |
| rl-6(t) |
rolled leaf-6 |
12%-lp-1 |
- |
547 |
| S-7 |
Hybrid sterility-7 |
9%-Rc |
KT |
577,579,596,597 |
| S-9 |
Hybrid sterility-9 |
19%-Est-l |
KT |
578 |
| se-2 |
photosensitivity-2 |
23%-g-1 |
- |
628 |
| spl-9 |
spotted leaf-9 |
triplo-7 |
IR |
137,401 |
| st-8 |
stripe-8 |
4cM-wp-l |
IR |
401 |
| Un-b |
Uneven grain |
18%-g-1 |
- |
548 |
| wp-1 |
white panicle-1 |
triplo-7 |
IR |
401 |
| Wph-l |
White-backed planthopper
resistance-1 |
near RG146B |
IR |
8,268,479,540 |
| xa-k* |
Xanthomonas campestris
pv. Oryzae resistance |
triplo-7 |
- |
232 |
| z-10 |
zebra-10 |
OcM-wp-l. trlplo-7 |
IR |
400,401 |
| Group 8 |
|
|
|
|
| Amp-2 |
Aminopeptidase-2 |
0 8.3cM to RG136 |
GI.IR |
120,369,589, 640 |
| Pi-11 (t)* (Pi-zh) |
Pyricularia oryzae
resistance-11 |
30 near RZ617 14.9cM
to Bpl27 |
IR |
540,640 |
| sug(su) |
sugary endosperm |
0 near XNpb397 |
KY |
95,134,142,174, 442,602 |
| v-8 |
virescent-8 |
115 near XNpb278 |
KY |
95,134,142,174 |
| Unlocated genes |
|
|
|
|
| Amp-4 |
Aminopeptidase-4 |
triplo-8 |
IR |
257,589 |
| Amy3D/E* |
Alpha-amylase-3D/E |
ll.lcM-RGI |
IR |
365,376,386 |
| An-4(t)* |
Awn-4 |
5.0%-TR7-8b |
RY |
427 |
| chl-8 |
chlorina-8 |
triplo-8 |
IR |
172,401 |
| chl-9 |
chlorina-9 |
triplo-6 |
IR |
172 |
| d-51(d-K-8) |
dwarf Kyushu-8 |
trisomic-8 |
KY |
134,142 |
| shr-2 |
shrunken endosperm-2 |
trisomic-8 |
KY |
263,439 |
| sk-2(t) (scl,fgr) |
scented kernel-2 |
6.7%-XNpbl26 2.1cM-RG28 |
NA,IR |
4,540,606 |
| ur-2 |
undulate rachis-2 |
trisomic-8 |
KY |
134,142 |
| xa(t)* |
Xanthomonascampestris
pv. Oryzae
resistance |
3.8cM-RG136 |
- |
xa(t)* |
| (Continued) |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| z-4 |
zebra-4 |
trisomic-8 |
KY |
134,142 |
| Group 9 |
|
|
|
|
| I-Bf |
Inhibitor for brown
furrows |
0 11.lcM to XNpb36 |
HK,KY |
92,94,131,134, 297,298,423 |
| lam(t) |
low amylose endosperm |
0 |
HK |
180 |
| Dn-1 |
Dense panicle-1 |
45 |
HK.KY |
298 |
| dp-2 |
depressed palea-2 |
45 3.3cM to XNpb385 |
HK.KY |
129,131,192,282 |
| drp-2 |
dripping-wet leaf-2 |
51 0cM to XNpb385 |
KY |
94,129,134,192 |
| Bp |
Burlush-like panicle |
63 1.4cM to XNpb339 |
KY |
131,134,192 |
| Unlocated genes |
|
|
|
|
| Adh-2* |
Alcohol dehydrogenase-2 |
trisomic-9 |
IR |
376 |
| Amy3A/B/C* |
Alpha-amylase-3A/B/C |
9.4cM-RZ228 |
IH |
365,376,386 |
| chs-l(t)* |
chlorosis caused by
low temperature (17ºC)-l |
27%-dp-2 |
KY |
34 |
| cyc-1* |
cyclophilin-1 |
4.6cM-RZ404 |
- |
540 |
| d-57(d(x)) |
dwarf |
21%-Dn-1 |
- |
83,610 |
| du-2120* |
dull endosperm (2120) |
triplo-9 |
IR |
166 |
| Est-3 |
Esterase-3 |
16.4%-XNpb257 |
NA.IR |
303,369,605 |
| Est-12 |
Esterase-12 |
29.1cM-RZ12 |
- |
19 |
| GIUP-I(GUP-I) |
Glutelinprecursor-I |
trisomic-9 |
KY |
450 |
| glup-2(gup-2) |
glutelin precursor-2 |
trisomic-9 |
KY |
450 |
| gm-1 (pd.sgm) |
gall midge resistance |
35%-I-Bf |
- |
423,424,460 |
| man |
Mannan in endosperm
cell wall |
4.3cM-XNpb293 |
NA |
607,608 |
| ms-10 |
male sterile-10 |
5%-Dn-l |
KT |
205,206,207 |
| R5s-3* |
5S ribosomal DNA-3 |
short arm |
NA |
33 |
| R45s-l*(rDNA-l) |
45s ribosomal DNA-1 |
1.0cM-XNpb315 |
NA |
51,265,266,540 |
| Group 10 |
|
|
|
|
| pgl |
pale green leaf |
0 3.5cM to XNpbl27 |
KY |
92,93,131,466, 617 |
| spl-10 |
spotted leaf-10 |
8 10.9cM to XNpbl27 |
KY |
92,93 |
| (Continued) |
| Gene |
Name |
Gene locus |
Institution 1) |
Literary No. |
| Rf-1 |
Pollen fertility restoration-1 |
12 7.5cM to XNpb291 |
RY.HK.NA |
53,432,465,466, 467,631 |
| fgl(fl) |
faded green leaf |
12.5 OcM to XNpb291 |
KY |
131,134,192,466, 605,617 |
| Ef-l.(=Ef-2) |
Earliness-1 |
28.5 |
RY.CH |
432.433,435, 558,561,569,571 |
| Unlocated genes |
|
|
|
|
| Bph-3 |
Brown planthopper
resistance-3 |
trisomic-10 |
NA.IR |
100,101,223,476 |
| bph-4 |
brown planthopper
resistance-4 |
30%-rk-2 near Bph-3 |
NA.IR |
100,101,223, 476,477 |
| du-l |
dull endosperm-l |
trisomic-10 |
KY |
134,442,443,604 |
| Ef(t)* |
Earliness introduced
from O. australiensis |
10.0cM-CD098 |
IR |
113 |
| Glh-3 |
Green leaf hopper
resistance-3 |
34%-bph-4 |
IR |
12,477,485 |
| glu-2(t)* |
lack of glutelin subunit-2 |
6.8cM-G1082 |
NA |
99 |
| rDNA-2* |
ribosomal DNA-2 |
0-llc M-XNpb32 |
NA |
33,51,265 |
| rk-2 |
round kernel-2 |
2.5%-TR10-11 7.5cM-pg1 |
KY |
134,617 |
| yg l |
yellow green leaf |
trlplo-10 |
IR |
228 |
| Group 11 |
|
|
|
|
| Pi-k |
Pyricularia orvzae
resistance-k |
0 17.8% to G181 |
NA |
64,155,202,203, 204,471 |
| d-27(d-t) |
bunketsuto tillering
dwarf |
32 2.6cM to pTA818 |
KY.HK |
1,132,134,138 |
| Xa-21* |
Xanthomonas campestris
pv oryzae resistance-21 |
37 2.2cM to pTA818 |
NA.IR |
1,102,176,387, 388,389 |
| z-2 drp-7 |
zebra-2 dripping-wet
leaf-7 |
46 0cM to CD0365 64 |
KY KY |
1,94,132,134, 138
445,447 |
| la |
lazy growth habit |
69 3.0cM to RG1094 |
HK.KY |
1,192,298 |
| v-4 |
virescent-4 |
80 |
KY |
136,138,358 |
| Pgd-1 |
Phosphogluconate dehydrogenase-1 |
85 near RG167 |
GI.IR |
119,120,122,123 369.540,689 |
| d-28(d-C) |
chokeldaikoku or long
stemroed dwarf |
95 |
KY |
134,138 |
| sp |
short panicle |
103 7.7cM to XNpb320 |
KY.HK |
130,134,138 |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| Adh-l |
Alcohol dehydrogenase-1 |
104 7. 3% to RG118 |
IR.GI |
1,16,117,122, 123,374,375,402 |
| Pr-a |
Pyricularia oryzae
resistance-a |
105 |
NA |
64,203,471 |
| sh-1 |
shattering-l |
108 |
HK |
298 |
| v-9 |
virescent-9 |
116 11.lcM to XNpbl83 |
KY |
94,446 |
| esp-2(rsp-2) |
endosperm storage
protein-2 |
124 |
KY |
211,212,215,440 |
| z-l |
zebra-1 |
132 |
KY |
131,132,134 |
| D-53(D-K-3) |
Dwarf Kyushu-3 |
141 |
KY |
134,136,138 |
| Unlocated genes |
|
|
|
|
| D-2 |
Dominant lethal from
O.sativa |
37.3%-Pgd-1. |
IR |
57 |
| esp-3(rsp-3) |
endosperm storage
protein-3 |
trisomic-11 |
KY |
212,214,215 |
| Fdp-l* |
Fructose-1, 6-diphosphatase-1 |
triplo-ll |
IR |
16,381 |
| gal (It) |
gametic letal |
24%-la |
KT |
537,553 |
| M-Pi-z |
Modifier for Pi-z |
11%-la |
YA |
60 |
| nal-2 |
narrow leaf-2 |
36%-la |
HK |
282 |
| PBR* |
Panicle blast resistance |
10.8cM-CD0226 |
NA |
46 |
| Pi-f |
Pyricularia oryzae
resistance-f |
15%-Pi-k |
NA |
471,555,633,634 |
| Pi-is-l(Rb-4) |
Pyricularia oryzae
resistance-is |
23%-la |
YA |
59 |
| Pi-kur-2* |
Pyricularia oryzae
resistance-kur-2 |
14%-la |
YA |
61 |
| Pi-se-l(Rb-l) |
Pyricularia oryzae
resistance-se |
9.5%-la |
YA |
62,63 |
| Pi-1 |
Pyricularia oryzae
resistance-1 |
4.7%-Pi-k 3.5cM-NpB181 |
IR |
110,237,271, 540 |
| Pi-7(t) |
Pyricularia oryzae
resistance-7 |
near RG16 |
IR |
582 |
| R5S-1* |
5S ribosomal DNA-1 |
near RG167 |
NA |
33,158,266,540 |
| S-3 |
Hybrid sterllity-3 |
1%-la |
Gl |
404 |
| Stv-b |
Stripe disease resistance |
3.0cM-XNpb257 |
NA |
70,584,585.586 |
| Xa-3 (Xa-w. Xa-4°Xa-6
Xa-9) |
Xanthomonas campestrispv.
oryzae resistance-3 |
22%-d-27 2.3cM-XNpbl81 |
IR |
43,325,326,327, 328,329,330,333
334,335,475,484 622,624 |
| Xa-4(Xa-4a) |
oryzae resistance-4 |
near Xa-3 1. 7cM-XNpb78 |
IR |
268,329,330,368 478,484,495,622
624,625,626 |
| Xa-10 |
Xanthomonaas campestris
pv oryzae resistance-10 |
27%-Xa-4 5.3cM-Oo72000 |
KY |
619,626 |
| 28%-la triplo-ll |
oryzae resistance |
28%-la triplo-ll |
|
232,629 |
| Gene |
Name |
Gene locus |
Institution 1) |
Literature No. |
| Xa-h* |
Xanthomonas campestris
pv oryzae resistance |
17%-Xa-a |
- |
629 |
| Group 12 |
|
|
|
|
| acp-1 |
Acid phosphatase-1 |
0 |
IR.GI |
42,65.116,117,118,120,122,123
369,589,640 |
| Acp-2 |
Acid phosphatase-2 |
0 1.9cM to RG181A |
IR.GI |
65,120,364,369, 589,640 |
| Pox-2 |
Peroxidase-2 |
26 |
GI.IR |
65,116,117,120, 121,122,123,
369,589 |
| nal-3(nal-2) |
narrow leaf-3 |
29 |
KY,HK |
131,134,192, 287,617 |
| d-33(d-B) |
bonsai to dwarf |
34 7.5cM to XNpbl48 |
KY |
131,134,192,287 |
| rl-3(rl-l) |
rolled leaf-3 |
36 near XNpb402 |
KY.IR |
95,116,131,134, 287,617 |
| spl-l(bl-12) |
spotted leaf-1 |
45 3.6cM to XNpb88 |
KY |
92,95,116,131, 134,137,192,617 |
| Sdh-l |
Shikimate dehydrogenase-1 |
49 4.9cM to XNpb402 |
IH.GI |
65,116,120,122,123,306,369,374
375,396,461 |
| Unlocated genes |
|
|
|
|
| Ald-3* |
Aldolase-3 |
near C1336 |
NA |
50 |
| Bph-l |
Brown planthopper
resistance-1 |
10.9cM-X Npb248 |
NA.IR |
100,101,476,647 |
| Bph(t)* |
Brown planthopper
resistance introduced from O. australiensis |
3.7cM-RG457 |
IR |
113,178 |
| du-4 |
dull endosperm-4 |
trisomic-12 31%-d-33 |
KY |
444,604 |
| ga-13 |
gametophytegene-13 |
10%-Acp-l |
CH |
384 |
| Hbv |
Hojablanca resistance |
|
IR |
178,554 |
| lec* |
Lectin |
near RG190 |
IR |
540 |
| Pi-ta (=Pi-4a) |
Pyricularia oryzae
resistance-ta |
5%-spl-1 0cM-XNpb289 |
NA.IR |
110,112,202,203237,271,278,392
452,471,582,630 |
| Pi-6(t)* |
Pyricularia oryzae
resistance-6 |
near RG81 |
IH |
269,540 |
| Pro-1* |
Prolamin-1 |
3.2%-XNpb88 |
KY |
441 |
| S-15 |
Hybrid Sterility-15 |
27%-Sdh-l |
KT |
27%-Sdh-l |
-
Name and address of institution are listed in appendix.
|
|
|
Appendix: Institution code
CH: Food Crops Research Institute, National Chung Hsing University,
Taichung, Taiwan
400, ROC
Gl: Genetic Stock Center, National Institute of Genetics, Mishima,
411 Japan. Fax
81-559-81-6804
HK: Plant Breedking Institute, Faculty of Agriculture, Hokkaido University,
Kita 9, Nishi
9, Kita-ku, Sapporo, 060 Japan, Fax 81-1 1-706-4934
IR: Rice Germplasm Center, International Rice Research Institute, P.
0. Box 933, Manila,
Pfilippines, Fax 63-2-817-8470 or 63-2-891-1292
KT: Plant Breeding Laboratory, Faculty of Agriculture, Kyoto University,
Kitashirakawa.
Sakyo-ku, Kyoto, 606 Japan, Fax 81-75-753-6025
KY: Plant Breeding Laboratory, Faculty of Agriculture, Kyushu University,
Hakozaki,
Fukuoka, 305 Japan, Fax 81-92-641-2928
NA:Gremplasm Center, National Institute of Agrobiological Resources,
Tsukuba
lbaraki-ken. 305 Japan, Fax 81-298-38-7408
NG: Laboratory of Plant Genetics and Breeding, Faculty of Agriculture,
Nagoy
University, Chikusa-ku, Nagoya, 464-01 Japan, Fax 81-52-781-4447
OK: Institute of Agricultural and Biological Science, Okayama University,
Kurashiki.
710 Japan, Fax 81 -864-21 -0699
RY: Plant Breeding Laboratory, College of Agriculture, University of
Ryukyus,
Nishihara. Okinawa, 903-01 Japan, Fax 81-9889-5-2864
YA: College of Agriculture, Yamagata University, Tsuruoka, 997 Japan,
Fax 81-235-28-2853



