Yo-Ichiro Sato1, Ling Hua Tang2
and Ikuo Nakamura3
1) Faculty of Agriculture, Shizuoka University,
Shizuoka, Japan
2) Jiangsu academy of agricultural sciences, Nanjing,
China
3) Iwate biotechnology research center, Kitakami,
Japan
We amplified fragments of chloroplast DNAs from charred rice grains excavated from Nabatake relics of Saga Prefecture (Japan, 2700 years old) and Caoxieshan relics of Jiangsu province (China, 6000 years old). DNA was extracted on a single-grain basis according to the method established by Nakamura and Sato (1991). DNA fragment was amplified by polimerase chain reaction (PCR) using two primers having sequences 5 '-tcg caa ccc ctt tcc gct aca c-3' (primer ORF) and 5'-tct tta gta atc cta cca ag-3' (primer ORF2) that are designed for amplification of a DNA fragment in ORF100 region at
Pst-12 site of rice chloroplast DNA. Polimerase chain reaction
was performed by 40 repeats of a 94°C(l min.)-51°C(30sec.)-72°C(l
min.) thermal cycle. Identification of DNA fragment amplified was attempted
by agarose-gel electrophoresis. Details for PCR and electrophoresis were
described in Nakamura and Sato (1991).
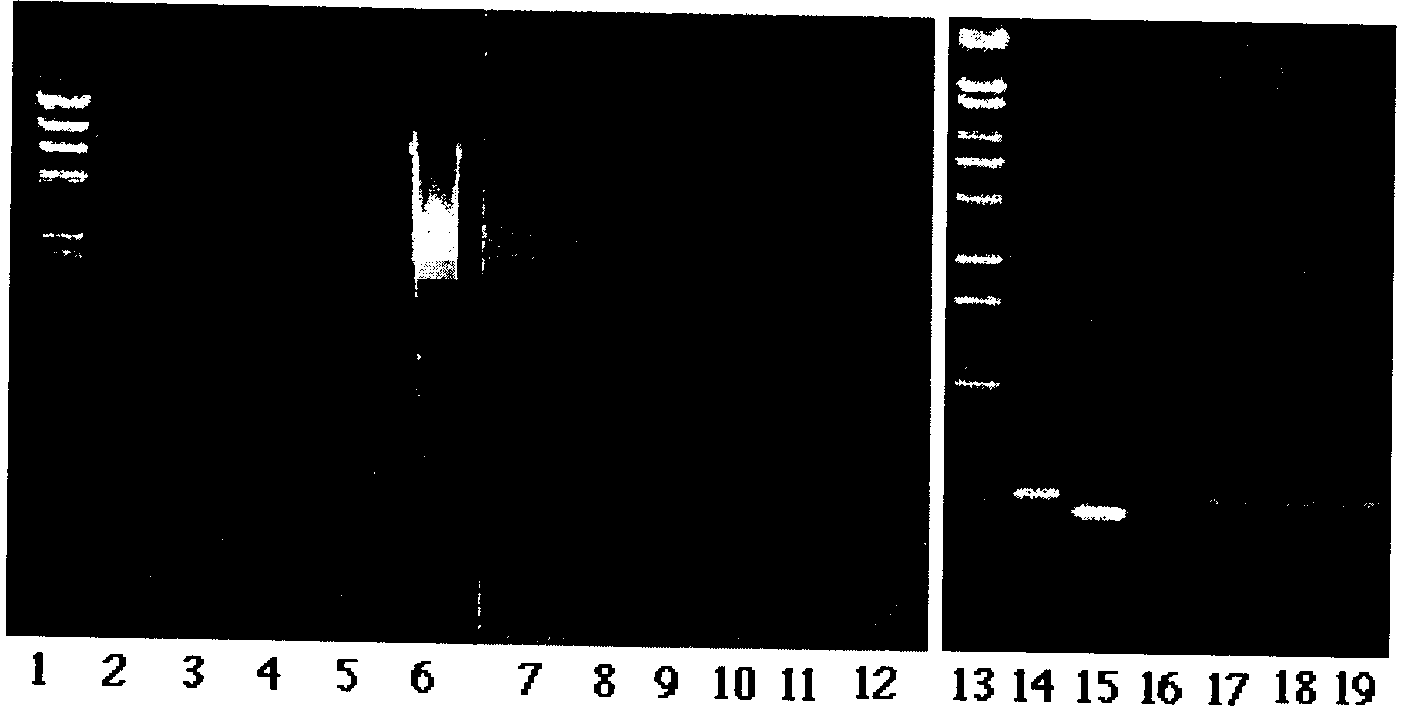
A 450 base-pairs fragment is amplified from more
than 85% of japonica cultivars. while a 69 base-pairs deletion exists
in more than 90% indica cultivars (Chen et al. 1993).
A 450 base-pairs DNA fragment was amplified from
charred grains excavated from both Nabatake and Caoxieshan relics. The
size of the fragment was as large as those amplified from the modern japonica
cultivars. A fragment of chloroplast DNA amplified from indica cultivars
was, on the other hand, as large as 380 base-pairs, that was 69 base-pairs
smaller than that of japonica cultivars (Chen et al. 1993).
To verify that the DNA fragment amplified is surely
rice DNA, southern-blot analysis was attempted. A PCR product from total
DNA extracted from a japonica cultivar Koshihikari was used as a
probe. It was shown that the DNA fragments amplified from Nabatake charred
grain properly hybridize with that from Koshihikari.
From these results, we suggest that the two charred
grains had japonica-type chloroplast DNAs. Predominant rice cultivars
in Japan and lower basin of Yangtze river were considered to be japonicas,
although nuclear DNA type and phenotypes were not analyzed yet.
References
Chen, W. B., I. Nakamura, Y. I. Sato and H. Nakai, 1993. Distribution
of deletion type in cpDNA of cultivated
and wild rice. Jpn. J. Genet.
68: 597-603.
Nakamura, I. and Y. I. Sato, 1991. Amplification of DNA fragments isolated
from a single seed of ancient rice
(AD800) by polymerase chain
reaction. Chinese J. Rice Sci. 5: 175-179.