43. RAPD and RFLP mapping of a submergence tolerance locus in rice
Kenong Xu and David J. Mackill
Department of Agronomy and
Range Science and USDA-ARS
University of California,
Davis, CA 95616, USA
Submergence tolerance is an important characterisitc
to breed into improved rice cultivars for rainfed lowland and deepwater
areas. It may also be useful as a strategy for weed management in areas
where rice is seeded directly into standing water, and deeper water levels
(20cm) can be used for weed suppression. Genetic studies have been conducted
(Mohanty et al. 1982; Haque et al. 1989), but the specific
genes controlling tolerance have not been identified.
We studied the inheritance of submergence tolerance
in a cross between the highly tolerant indica line IR40931-26-3-3-5 and
the susceptible japonica line P1543851. IR40931-26 has a similar level
of tolerance to FR13A, from which it derived the trait (Mackill et al.
1993). At 8 DAS, F3 families were subjected to 14-16 d submergence
in greenhouse tanks. Individual plants were scored on a scale of 1 (tolerance)
to 9 (susceptible) 1 wk after the water was removed.
Differences among F3 families were highly
significant, and means ranged from 1.6 to 8.9, compared to 1.5 and 8.4
for the tolerant and susceptible families. DNA from the F2 plants
giving rise to the nine most tolerant and most susceptible F2 families
were bulked and assayed with 624 RAPD markers. Five RAPD bands associated
with either the tolerant or susceptible families were asseyed on the entire
F2 population. These five bands were linked to each other. RFLP
markers from Cornell University, USA (McCouch et al. 1988), and
from the Rice Genome Program, STAFF Institute, Tsukuba, Japan (Kurata et
al. 1994) were used to map these RAPD markers to the end of chromosome
9 (Fig. 1). Analysis by Mapmaker/QTL (Lincoln et al. 1992) identified
a locus near the RFLP
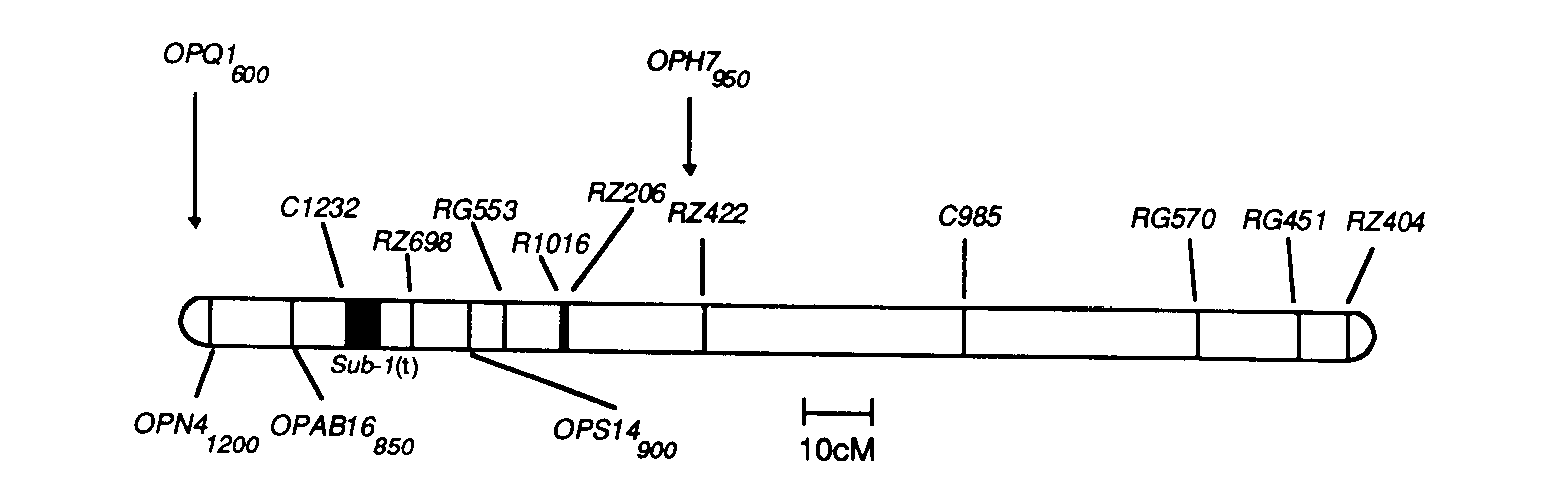
Fig. 1. RFLP map of rice chromosome 9 showing RAPD and RFLP markers
and putative location of Sub-1(t) (dark bar) based on Mapmaker/QTL
analysis.
marker C1232 responsible for 69% of the phenotypic variance for this trait.
An additional 79 markers distributed on the other rice chromosomes did
not detect QTLs. It is not clear whether this locus represents a single
gene, or closely linked genes or loci. We have tentatively designated this
locus as Sub-l(t). Identification of molecular markers closely linked
to Sub-1(t) should greatly facilitate the transfer of submergence
tolerance into improved lines.
Reference
Haque, Q. A., D. HilleRisLambers, N. M. Tepora and Q. D. dela Cruz,
1989. Inheritance of submergence
tolerance in rice. Euphytica
41: 247-251.
Kurata, N., Y. Nagamura, K. Yamamoto, Y. Harushima, N. Sue, J. Wu,
B. A. Antonio, A. Shomura, T. Shimizu,
S. Y. Lin, T. Inoue,
A. Pukuda, T. Shimano, Y. Kuboki, T. Toyama, Y. Miyamoto, T. Kirihara,
K. Hayasaka,
A. Miyao, L. Monna, H. S.
Zhong, Y. Tamura, Z. X. Wang, T. Momma, Y. Umehara, M. Yano, T. Sasaki
and Y. Minobe, 1994. A 300
kilobase interval genetic map of rice including 883 expressed sequences.
Nature Genetics 8:
365-372.
Lincoln, S., M. Daly and E. Lander, 1992. Mapping genes controlling
quantitative traits with
MAPMAKER/QTL 1.1 No. Whitehead
Institute Technical Report, 2nd ed.
Mackill, D. J., M. M. Amante, B. S. Vergara and S. Sarkarung, 1993.
Improved semidwarf rice lines with
tolerance to submergence
of seedlings. Crop Sci. 33: 749-753.
McCouch, S. R., G. Kochert, Z. H. Yu, Z. Y. Wang, G. S. Khush, W. R.
Coffman and S. D. Tanksley, 1988.
Molecular mapping of rice
chromosomes. Theor. Appl. Genet. 76: 815-829.
Mohanty, H. K., B. Suprihatno, G. S. Khush, W. R. Coffman and B. S.
Vergara, 1982. Inheritance of submergence
tolerance in deepwater rice.
p. 121-134, In Proceedings of the 1981 International Deepwater Rice
Workshop. Int. Rice Res.
Inst., Los Banos, Philippines.

