35. Identification of RAPD markers linked to rice thermosensitive
genetic male
sterility gene by bulk
segregant analysis
P. K. Subudhi, Reena P. Borkakati, S. S. Virmani and N. Hung
Genome Mapping Laboratory, Plant Breeding, Genetics and Biochemistry
Division, IRRI. Philippines
Exploitation of heterosis through production of hybrids
is a major approach to increase productivity in rice. Usually the hybrid
seed production is carried out by the 3-line breeding system utilising
cytoplasmic genetic male sterility, which is the major limiting factor
for large scale production of hybrid seeds in tropics and subtropics. The
discovery and application of thermosensitive genetic male sterility (TGMS)
is likely to have great impact in those areas because of many advantages
in its use compared to CMS system such as easier multiplication of TGMS
seeds, no need of maintainers for establishing male sterile line, no need
of restorer parents etc. The cytoplasmic base can also be diversified,
since TGMS genes can be incorporated into any rice cultivar to use it as
female parent of hybrids. The incorporation of TGMS gene through conventional
breeding procedure is quite cumbersome, it involves identification of TGMS
plants in segregating generations and induction of seed fertility by ratooning
under appropriate fertility inducing temperature regime. Mapping of this
gene using molecular markers should help in exercising marker-aided selection
for transferring the TGMS gene to different genetic background.
A TGMS mutant line IR32364TGMS developed through
mutagenesis at IRRI was used, which showed complete male sterility at day/night
temperature of 32°/24°C, but partial fertility at 27°/21°C
and 24°/18°C in IRRI phytotron (IRRI 1992; Virmani and Voc 1991).
A mapping population was developed from the cross IR32364TGMS X IR68. The
genetic analysis indicated this trait to be under control of a single recessive
gene. The individuals in F2 generation were phenotyped and progenies of
fertile F2 individuals were grown to distinguish homozygous
and heterozygous genotypes. Single plants of these population were evaluated
for pollen fertility (determined by using 1 % IKI solution) and spikelet
fertility (determined by using two bagged panicles per plant). Individuals
with less than 20% pollen fertility were grouped under sterile class and
partial fertile individuals were excluded from analysis.
DNA of two parents and 64 F2 individuals
was isolated following the procedure of Dellaporta et a/.(1983)
and the bulk segregant analysis (Michelmore et al. 1991) was performed
in conjuction with RAPD (Random Amplified Polymorphic DNA) technique. Equal
amounts of DNA from 10 completely sterile and 10 fully fertile F2
individuals were pooled to constitute the sterile and fertile bulks respectively.
The two bulked samples along with parents were then subjected to RAPD analysis
using 10-mer random primers obtained from Operon Technologies Inc. Alameda,
California. The amplification reaction condition of Williams et al.
(1990) was followed with minor modification in Techne thermocycler. Amplification
profile was 94°C for 2 min followed by 45 cycles of 1 min at 94°C,
1 min at 36°C, 2 min at 72°C with a final extension time of 7 min
at 72°C. Amplification products were analysed in 1.5% ethidium bromide
containing agarose gel.
After surveying 389 random primers, five RAPD markers
generated by five primers; OPF18, OPB19, OPAC1, OPAC3 and OPAA7 were found
to be putatively linked to TGMS gene. Further analysis with all 34 homozygous
sterile and homozygous fertile F2 individuals showed cosegregation
of these markers with the trait. Fig. 1 shows a typical result of the bulk
segregant analysis with one of the primers. Out of 5 markers, four were
sterility specific (OPE 18-2600, OPB 19-750, OPAC1-300, OPAA7-550) and
one fertility specific (OPAC3-640). All the polymophic fragments were of
multicopy sequences except OPF18-2600. The OPF1 8-2600 fragment when used
as probe also showed cosegregation with TGMS in Southern blot analysis
of progenies. Preliminary analysis revealed linkage of all these markers
to the TGMS gene flanking on both the sides (Fig. 2). These fragments are
now being cloned. Mapping and sequencing of these clones are underway,
which can be helpful for marker assisted selection, while transferring
to different genetic background. Recently a TGMS gene has been reported
to be mapped on chromosome 8 (Wang et al. 1995), but we could not
detect linkage with any of the marker from this chromosome thus indicating
that the mutant gene of IRRI line may be different.
We gratefully acknowledge the technical help rendered
by Arnold Parco for this work. PKS was supported with a post-doctoral fellowship
by the Rockefeller Foundation.
 Fig. 1. DNA amplification pattern obtained with random primer OPF18
in bulked segregant analysis. The polymorphic fragment of 2600 bp is indicated
by an arrow. M. Marker, SP. Sterile parent, IR32364TGMS, FP. Fertile parent
IR68, FB. Fertile bulk, SB. Sterile bulk.
Fig. 1. DNA amplification pattern obtained with random primer OPF18
in bulked segregant analysis. The polymorphic fragment of 2600 bp is indicated
by an arrow. M. Marker, SP. Sterile parent, IR32364TGMS, FP. Fertile parent
IR68, FB. Fertile bulk, SB. Sterile bulk.
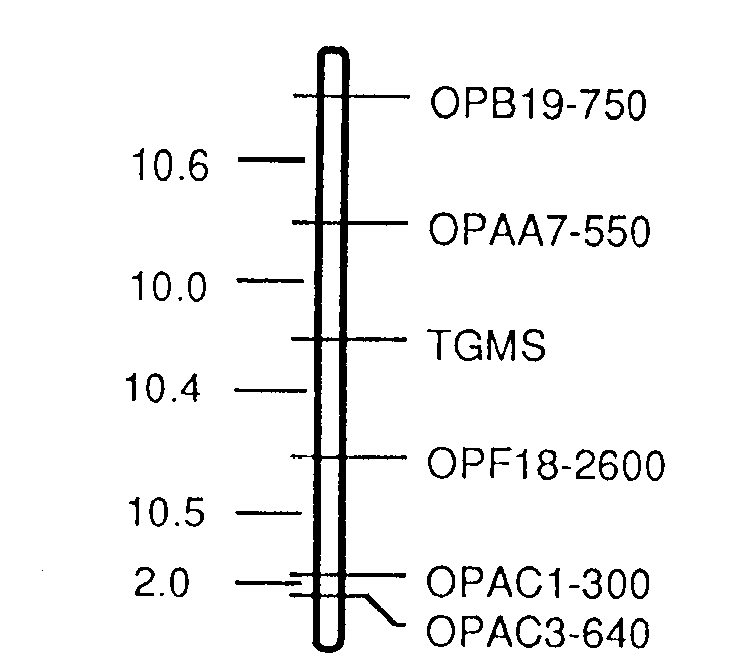 Fig. 2. RAPD map of the region surrounding the gene for thermosensitive
genetic male sterility (TGMS), based on segregation data from the cross
IR32364TGMS X IR68. Distances(cM) are given on the left side and the markers
are on the right side of the vertical line.
Fig. 2. RAPD map of the region surrounding the gene for thermosensitive
genetic male sterility (TGMS), based on segregation data from the cross
IR32364TGMS X IR68. Distances(cM) are given on the left side and the markers
are on the right side of the vertical line.
References
Dellaporta, S. L., J. Wood and J. B. Hick, 1983. A plant DNA minipreparation:
Version II. Plant Mol. Biol.
Report 1:19-21.
International Rice Research Institute, 1992. Annual Report for 1991,
IRRI, P. 0. Box 933. Manila, Philippines.
Michelmore, R. W., I. Paran and R. V. Kesseli, 1991. Identification
of markers linked to disease resistance genes
by bulk segregant analysis:
a rapid method to detect markers in specific genomic regions using segregating
populations. Proc. Natl.
Acad. Sci. USA 88: 9828-9832.
Virmani, S. S. and P. C. Voc, 1991. Induction of photo and thermo-sensitive
male sterility in Indica rice. Agron.
Abst. 119.
Wang, B., J. Wang, W. Xu and W. Wu, 1995. Tagging and mapping of rice
thermosensitive genic male sterile
(TGMS) gene. Plant Genome
III, Proceedings of the International Conference on the status of plant
genome
reseach, Jan. 15-19, 1995,
San Diego, California, pp.92. (Abstract)
Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski and
S. V. Tingey, 1990. DNA polymorphisms
amplified by arbitrary primers
are useful as genetic markers. Nucleic Acids Res. 18:6531 -6535.


