Zi-Xuan Wang, Akio Kojima1,
Nori Kurata, Yuzo Minobe and Takuji Sasaki
Rice Genome Research Program,
National Institute of Agrobiological Resources, 2-1-2 Kannondai.
Tsukuba 305, Japan / STAFF
(Society for Techno-innovation of Agriculture, Forestry and Fisheries)
Institute, 446-Ippaizuka,
Kamiyokoba, Tsukuba, 305, Japan
1) Present address: Kyushu
National Agricultural Experimental Station, Kirishima 5-2, Miyazaki 880,
Japan
In our rice genome mapping project, we could construct
a high-density linkage map by using molecular markers (Kurata et al.
1994). However, there were a number of genomic clones that could not be
assigned to respective chromosomes, because of lack of RFLPs between the
parent varieties using the given enzymes. Those clones are likely to contain
a genomic fragment with a specific function and a specific chromosomal
position. We are interested in assigning those clones to their respective
chromosomes. We describe here a method to assign the non-RFLP producing
genomic clones rapidly by using rice aneuhaploids, tetrasomics and trisomics.
Three types of aneuhaploid (Aneuhaplo 5, 9 and 11,
2n=13), 4 types of tetrasomics (Tetra 6, 7, 10 and 12; 2n=26) and 4 types
of trisomics (Triple 1, 2, 4 and 8; 2n=25), which cover 11 chromosomes
of rice (Iwata and Omura 1984; Wang and Iwata 1995), were used to assign
clones to specific chromosomes. The aneuploid for rice chromosome 3 was
not available in the experiment. Assignments of DNA clones to specific
chromosomes were performed by dosage comparison among genomic Southern
hybridization of all aneuploids. These aneuploids were in the background
of a japonica variety of "Nipponbare". The diploid "Nipponbare"
was used as a control. Eleven aneuploids and "Nipponbare" were grown in
the greenhouse. DNA extraction and Southern blots were performed according
to Kurata et al. (1994).
A total of 33 genomic clones from a Pst I
library (pNP, 11 clones) and a Hind III library (pNH, 22 clones),
made using a japonica variety "Nipponbare" and Bluesript II SK+
vector, were used for chromosome assignment. All these clones were single
copy in the genome and showed monomorphism in Southern analysis between
the two parent varieties "Nipponbare" and "Kasalath" with the 8 enzymes:
Apa
I, BamH I, Bgl II, Dra I, EcoR I, EcoR V, Hind
III and Kpn I used in our mapping project (Kurata et al. 1994).
Chromosomal assignment of clones was determined
by using the dosage effects of the extra chromosomes of aneuploids, as
has been described in tomato (Young el al. 1987) and in rice (Wang
et
al. 1995, Yu et al. 1995). In such an analysis, the most probable
source of error is variation in the quantity of DNAs among aneuploids loaded
on individual lanes of the gel used for the Southern analysis. To avoid
this problem, another clone having a different size was used together in
the hybridization experiment as an internal control probe of each lane.
This could help adjust signal intensities of each lane and made it possible
to confirm an increased hybridization signal of a clone in a given aneuploid.
Two clones having different sizes were mixed and digoxigenin-labeled to
hybridize to DNAs from aneuhaploids, tetrasomic, trisomics and "Nipponbare".
As shown in Figs. 1 and 2, hybridization signals obtained as autoradiograms
could clearly show which aneuploid had an extra copy or copies of the cloned
DNA. By looking at the increase in dosage of a DNA clone in one of the
aneuhaploids, tetrasomics, or trisomics, the chromosomal location of the
DNA clone was determined.
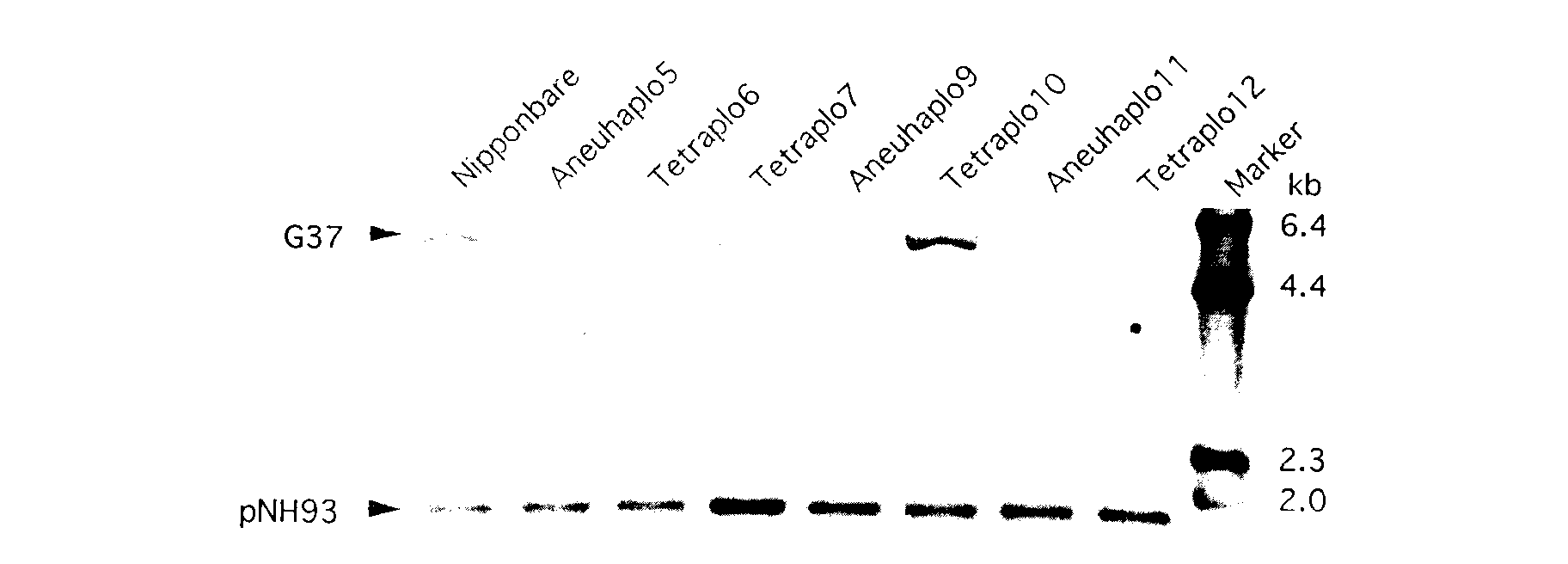
Fig. 1 shows an autoradiogram of two clones, pNH93
and G37, probed onto Hind III-digested DNAs from 3 aneuhaploids,
4 tetrasomics and "Nipponbare". pNH93 could be easily assigned on chromosome
7 and G37 on chromosome 10, as indicated by the increased signals in the
DNAs from Tetra7 and Tetra10, respectively. In this case, both clones hybridized
to the tetrasomics with high intensity signals were unequivocally determined
to be located on these chromosomes. G37 was used as an internal control.
The assignment of G37 to chromosome 10 determined in this study consistent
with the previous mapping data reported by Saito et al. (1991).
Because aneuhaploids and tetrasomics only cover
7 rice chromosomes, we also used 4 types trisomics to increase the coverage
of chromosomes and to facilitate the effectiveness of the assignment. Fig.
2 shows an autoradiogram of two clones, pNH 146 and pNH221, probed onto
Hind III-digested DNAs from 3 aneuhaploids, 4 tetrasomics, 4 trisomics
and "Nipponbare". The clone pNH146 was also easily assigned to chromosome
1, as indicated by the increased signals in the DNA from Triple 1. pNH221
could not be assigned, because of lack of dosage effect in the aneuploids
used in the experment. This clone may belong to chromosome 3, which is
only missing chromosome in this study. We found trisomics also useful in
this kind of analysis for dosage comparison. However, when trisomics were
used alone in the analysis, it is difficult to determine the dosage effect
with certainty, since the difference in 3 versus 2 dosages is not so great.
By using rice aneuhaploids and tetrasomics, it becomes easier to determine
the dosage effect, because the dosage difference is 4 versus 2 in tetrasomics
and 2 versus 1 in aneuhaploids. This is easily seen by comparing the signal
differences between Fig. 1 and Fig, 2. Aneuhaploids and tetrasomics are
thus more effective than trisomics in this kind of studies.
Out of the 33 clones tested, we have clearly assigned
24 clones to 8 rice chromosomes (Table 1). Among them, a few clones could
be mapped on our high-density
Table 1. Chromosomal assignment of Pstl (pNP) and Hind
III (pNH) clones
| Chromosome | Clones |
| 1 | pNP42, pNH6, pNH69, pNH 104, pNH 146, pNH 166 |
| 2 | pNP 101, pNH21, pNH72, pNH88 |
| 5 | pNH32 |
| 6 | pNP63, pNP25, pNH77 |
| 7 | pNH68, pNH93, pNH129, pNH139, pNH263 |
| 9 | pNP 103 |
| 10 | pNP 167, pNH84, pNH245 |
| 12 | pNH 123 |

We are grateful to Professor N. Iwata. Plant Breeding Laboratory, Faculty of Agriculture, Kyushu University for supporting this work, and Dr. 1. Havukkala, STAFF Institute, for comments on the manuscript.