M. Eigluchi1 and
Y. Sano2
1) National Institute of
Genetics, Mishima. 411 Japan
2) Plant Breeding Institute,
Faculty of Agriculture, Hokkaido University, Sapporo, 060 Japan
| Gene
combination
(A) (B) |
No. of phenotypes | Recombination value (sd) | X2
(df=3) |
||||||
| AB | Ab | aB | ab | Total | |||||
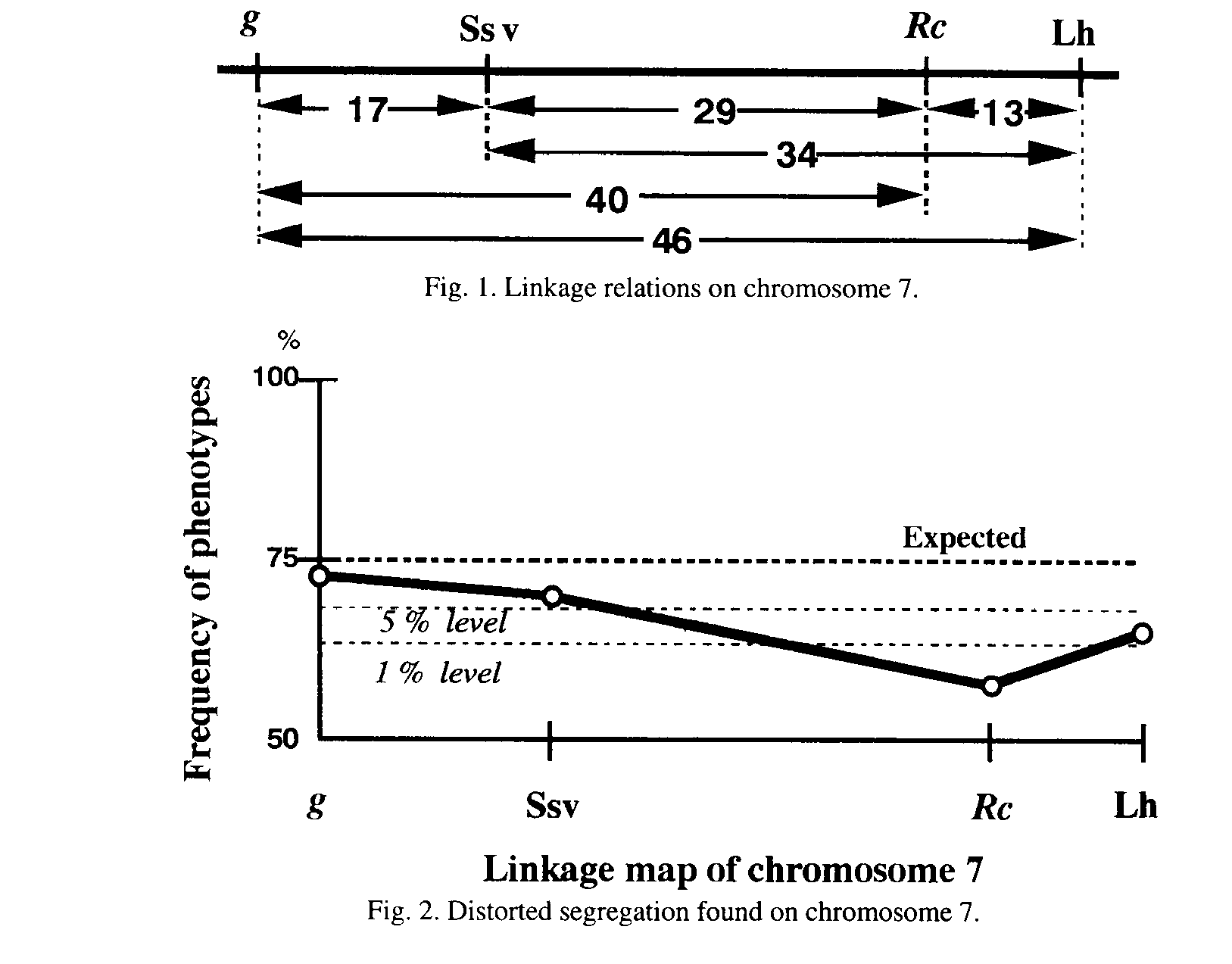
| Ssv(t) | g | Obs. | 84 | 5 | 10 | 31 | 130 | 0.17(0.04) | 6.0 ns |
| Exp. | 87.4 | 10.1 | 10.1 | 22.4 | |||||
| Ssv(t) | Rc | Obs. | 68 | 21 | 11 | 30 | 130 | 0.29(0.05) | 16.6** |
| Exp. | 81.4 | 16.1 | 16.1 | 16.4 | |||||
| Lh(t) | Rc | Obs. | 75 | 10 | 4 | 41 | 130 | 0.13(0.03) | 15.8** |
| Exp. | 89.6 | 7.9 | 7.9 | 24.6 | |||||
| Ssv(t) | Lh(t) | Obs. | 70 | 19 | 15 | 26 | 130 | 0.34(0.05) | 11.5** |
| Exp. | 79.2 | 18.3 | 18.3 | 14.2 | |||||
| Rc | g | Obs. | 66 | 13 | 28 | 23 | 130 | 0.40(0.06) | 17.8** |
| Exp. | 76.7 | 20.8 | 20.8 | 11.7 | 6.0 ns | ||||
| Lh(t) | g | Obs. | 67 | 18 | 27 | 18 | 130 | 0.46(0.06) | 6.0* |
| g | 74.5 | 23.0 | 23.0 | 9.5 | |||||
Note; The genotype of F1 was g+ Ssv(t) Rc Lh(t)/g
Ssv(t)+ rcLh(t)+.
ns shows non-significance.
* and ** show significance at 5% and 1%, respecitvely. Fig. 2,
suggesting that the introduced segment is quickly eliminated when backcrossed
with T65glg.