F. Research Notes
I. Wild species, evolution and chromosome studies
1. Phylogenetic study of A-genome species of genus Oryza using
nuclear RFLP
Kazuyuki Doi1,
Atsushi Yoshimura1, Mutsuko Nakano1, Nobuo Iwata1
and Duncan A. Vaughan2
2) National Institute of Agrobiological Resources. Tsukuba 305, Japan
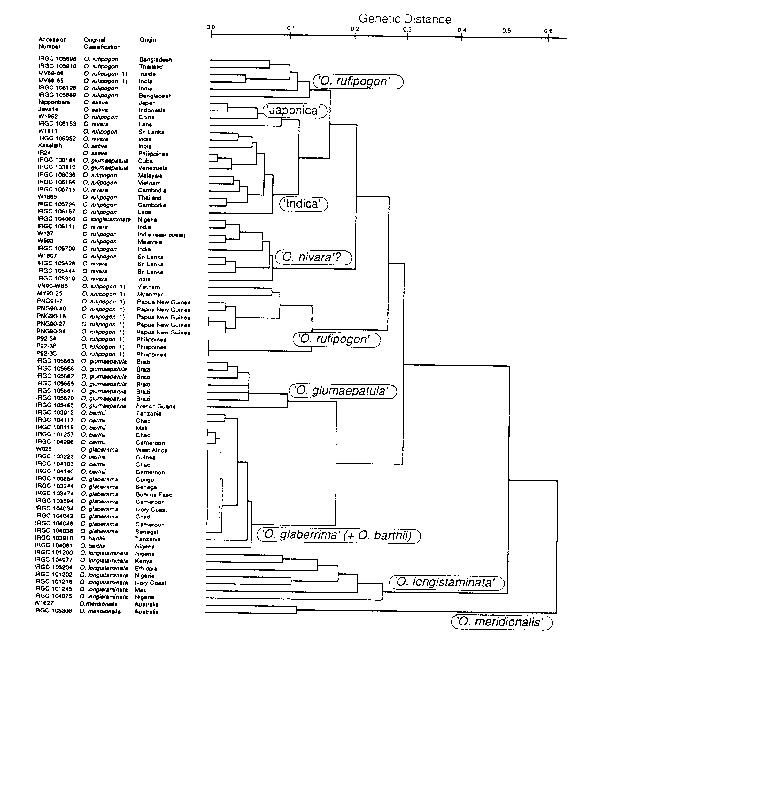
A dendrogram constructed by using 76 accessions including 12 accessions previously analyzed is shown in Fig. 1. A-genome species were classified into major groups. Asian group (0. sativa. 0. rufipogon and (O. nivara). 0. glumaepatula group, 0. glaberrima + 0. barthii group, 0. longistaminata group and 0. meridionalis group. 0. glumaepatula had closer affinity with 0. glaberrima + 0. barthii group than with Asian 0. rufipogon. although they had been considered to be a subtype of 0. rufipogon. Except for this. relationship among species matched well with previous studies.
Asian group which contains 0. sativa, 0. rufipogon and 0. nivara consisted of loosely knitted four or five groups, each corresponding to Indica (including some (9. rufipogon), Japonica (including some 0. rufipogon), 0. rufipogon (consisting of several groups) and 0. nivara (Fig. 1). There is a need for further analysis to determine the relationships of these sub-groups.
Three accessions of perennial 0. rufipogon from Papua New Guinea (excluded from Fig. 1 ) carried both fragments of (O. rufipogon and 0. meridionalis, suggesting the occurrence of natural hybridization between the species.
References
Nakano, M.. A. Yoshimura and N. lwata. 1992. Phylogenetic study of cultivated
rice and its wild relatives by
RFLP. Rice Genet. Newslett.
9: 132-134.
Nei. M.. 1990. Molecular Evolutionary Genetics. BAIFUKAN. Tokyo. (Japanese)
Saito, A.. M. Yano, N. Kishimoto, M. Nakagahra, A. Yoshimura, K. Saito,
S. Kuhara, Y. Ukai, M. Kawase, T. Nagamine, S.
Yoshimura, O. Ideta, R.
Ohsawa, Y. Hayano, N. Iwata and M. Sugiuru. 1991. Linkage map of restriction
fragment length
polymorphism loci
in rice. Japan. J. Breed. 41: 665-670.
Sokal, R. R. and C. D. Michener. 1958. A statistical method for evaluating
systematic relationships. Sci. Bull..
University of Kansas 38:
1409-1438.