I. Proposition of rules for gene symbolization in rice
As a result of new developments in rice genetics during the last two decades, the need for re-examination and revision of the gene symbols and the nomenclatorial rules has become evident.Realizing this, the Committee on Gene Symbolization, Nomenclature and Linkage has established new rules for gene symbols in rice. In principle, these rules follow the international rules adopted earlier by the International Rice Commission (IRC).
Rules for gene symbols in rice
1. Chromosomes. The chromosomes are assigned Arabic numerals in descending order of their pachytene length (or cenrtomere position in case of ambiguity of length). Short arms are symbolized by "S", long ones by "L" (Example:1S,1L).
2. Lingage groups.Linkage groups are assigned numerals corresponding to the respective chromosomes. As soon as the arm location of a gene is known, the gene locus numbering will be revised to reflect that information. The zero position will be assigned to the gene located on the distal end of the short arm.
3. Genes. (1) In naming of genes, the use of international language (English) is preferred. Genes should be named only after reasonable evidence has been obtained for their existence. (2) Gene symbols derived from names which describe the character modifications should be written in Italics and be as short as two or three letters. However, the symbols which have been commonly used by many workers in the past should be retained even if they do not fit this rule Example: C(Chromogen); A(Activator) and wx(glutinous endosperm). (3) Whenever unambiguous, the name and symbol of a dominant gene begin with a capital letter, and the recessive with a small letter. If the two alleles are equivalent or codominant or if their dominance relations are not consistent, capitalized symbols may be assigned to one of the alleles at the author's discretion.(4) Standard or wild type allels are designated by a gene symbol with "+" as a superscript. When it is clear in the text which gene is being discussed, the normal allele may be designated simply by the "+" symbol (Example: +, al+.) The standard strain may be any strain selected by the worker as long as the strain being used and its genetic formulae are made explicit. (5) Multiple alleles at the same locus are designated by appropriate letter or number superscripts (Example: d-18+, d-18k, d-18h, Pgi-11, Pgi-12). (6) The genes that are non-allelic but are indistinguishable from each other phenotypically (including duplicate and complementary genes) are designated by the same base letter but are differentiated by number or letter subscript. The number and letters may be written on the same line with those after hypen indicating the gene loci (Example: d-1, d-2, d-3, or D\1\, d\2\, d\3\, Hl-a Hl-b). (7) Inhibitors, suppressors, enhancers and modifiers are designated by the symbols, I, Su, En and M or by i, su, en and m if they are recessive, followed by a hyphen and symbol of the gene affected (Example: I-Pl- 1, Su-g\1\) (8) When a new gene is identified but its allelic relationships with previously reported genes with similar effects are not known, it is denoted by adding (t) to its symbol (t means tentative) (Example: d-50(t)). The suffix (t) is deleted when the allelic relationship is determined.
4. Structural change of chromosomes. Chromosomal changes are denoted by a symbol showing the type of aberration plus the chromosome number(s) involved. The symbols used are: Dp for duplication, In for inversion, T for translocation, Tp for transposition. To distinguish between the similar aberrations involving the same chromosome(s), lower-case letters are used following the chromosome numbers (Example: In(1)a In(1)b, T(1-2)a T(1-2)b).
5. Aneuploids. Monosomics and primary trisomics are designated according to the additional chromosome (Example: Mono-1, Mono-2, Triplo-1, Triplo-2).
6. Cytoplasms. Names and symbols of cytoplasmic genes follow the rules for nuclear genes. The symbol and the abbreviated name of species or variety enclosed within brackets are preceded by the genic formulae. (Example: (ms- bo)).
7. Priority and validity of symbols. (1) Priority of the gene nomenclature will be protected if the symbol is in accordance with these rules. (2) Inheritance data and information on the gene stocks (preferably with a photograph of mutant) should be presented to the convener of the Committee on Gene Symbolization, Nomenclature and Linkage Groups requesting the validation of the symbol.After an examination of priority, the convener will notify the author of recognition of the symbol and include the symbol in the list of gene symbols to be published in the Rice Genetics Newsletter. Publication of a research note on all the new mutants in the Rice Genetics Newsletter is recommended. (3) Viable seeds of the new gene stocks should be maintained by the respective authors. Seed samples should be sent to the Rice Genetics Stock Center.
8. Amendments. Amendments of the rules can be suggested by the Committee on Gene Symbolization, Nomenclature and Linkage Groups when necessary, and will come into effect when approved by the Co-ordinating Committee of the Rice Genetics Cooperative. Amendments will be announced in the Rice Genetics Newsletter.
II. Appointment of coordinators for rice gene symbols
On the occasion of the meeting of the Coordinating Committee of RGC held in Bangkok on November 26, 1985, Dr. Oka proposed the assignment of coordinators for monitoring rice gene symbols. In consultation with the members of the Committee, the following coordinators have been appointed.
Traits-Coordinators
===================
Chlorophyll and leaf spots - N. Iwata
Dwarfish - Y. Futsuhara, H. Kikuchi and J.N. Rutger
Heading behavior - S. Sato, H. Yamagata and K.H. Tsai
Leaf and culm - K. Mori and O. Kamijima
Endosperm - H. Satoh and E. Amano
Coloration - T. Kinoshita
Disease resistance - T. Ogawa and K. Toriyama
Insect resistance - G.S. Khush and R. Ikeda
Male sterility - C. Shinjo and S.S. Virmani
Panicle and spikelet - K. Takeda and G.S. Khush
Isozymes - H. Morishima and J.C. Glaszmann
Gametic effect and sterility - M. Nakagahra, Y. Sano and Y.I. Sato
Coordinators will have the following responsibilities:
1. To maintain current information and serve as a resource person for that trait.
2. To monitor ans assist with assigning new gene symbols.
3. To re-examine and revise the tentative gene symbols which were enumerated in RGN, Vol.1 and to conduct the allelism tests.
4. To prepare an annual report for the RGN.
III. Reports from the coordinators
1. Gene Symbols for chlorophyll deficiencies and leaf spots Nobuo Iwata, Faculty of Agriculture, Kyushu University, Higashi-ku, Fukuoka, 812 Japan
Following gene symbols for chlorophyll deficient mutants were proposed in RGN1.
al: albino(lethal white)
chl: Chlorina
st: striped
v: virescent
z: zebra
For the xantha (lethal yellow), the use of l-y, xa and x have been proposed by the Japanese Interim committee but no agreement has been reached as yet. The physiological leaf spots (non-pathogenic leaf spot mutant) are considered to be partly overlapping with the chlorophyll deficient mutants in a broad sense, but the committee has not reached agreement on standard gene symbols as yet.
Historically, Jones (1952) first reported two physiological disease mutants. One of these was controlled by an incompletely recessive gene and the other by a recessive gene. However, Jones did not assign gene symbols. Jodon (1957) described a black leaf spot occurring in the variety Magnolia and assigned the gene symbol lk to this recessive trait.
In 1959, IRC (international Rice Commission) recommended the use of a gene symbol bl for the physiological leaf spots on the basis of character expression of a dark brown or blackish mottled discoloration of leaf. Therefore, bl-1, bl- 2 and bl-3 were assigned for the mutants described by Jones (1952) and Jodon (1957), respectively.
Nagao and Takahashi (1954) used mt for the mottled glumes and leaves but later they changed the symbol to bl-1, conforming to the IRC rules (Nagao and Takahashi 1963). Takahashi et al. (1968) described four recessive genes, bl-3, bl-4, bl-5, bl-6, for the brown spots or speckles. In addition, a single recessive genes, ysl-1, was described for the yellow spots on leaves (Takahashi et al. 1968).
Iwata et al. (1978) assigned spl gene symbols to several kinds of physiological leaf spot mutants including not only blackish or brown but also reddish or other mottled colors, and nine recessive genes, spl-1 to spl-9 were reported.
Phenotypic description of these genes are as follows:
spl-1: Partial discoloration of leaves and stems.
spl-3, spl-5 and spl-7: Relatively small reddish brown spots scattered over the whole surface of leaves.
spl-4 and spl-6: Relatively large brown spots scattered on the leaves, but not so numerous as in spl-3, spl-5 and spl-7.
spl-8: Fine striped spots of reddish brown on whole surface of leaves.
spl-9: Small blackish brown spots on leaves and stems, but smaller in size. It should be noted that large or small reddish brown spots are present in these mutants.
As may be seen, two series of gene symbols, bl and spl, have been used in the
past for the physiological leaf spot mutants. Although bl gene symbol has a
priority over spl, many leaf spots show color expression such as red and yellow
which are different from black or brown spots to which bl gene symbol was
assigned. The use of spl is independent of spot color and is thus more
appropriate for the various leaf spots. In addition, it is known that the
manifestation of the leaf spot (e.g. color, size, shape, distribution and stage
of expression) is modified by the environmental conditions and/or genetic
background. Thus, it might be difficult to reach a definite conclusion on the
gene identification from the literature. Therefore, before you assign the new
gene symbols, please consult with the coordinator.
References
Iwata, N., T. Omura and H. Satoh, 1978. Linkage studies in rice (Oryza sativa L.). On some mutants for physiological leaf spots. J. Fac. Agr. Kyushu Univ. 22: 243-251.
Jodon, N.E., 1957. Inheritance of some of the more striking characters in rice. J. Hered. 48: 181-192.
Jones, J.W., 1952. Inheritance of natural and induced mutations in Caloro rice and observatons on sterile Caloro types. J. Hered. 43: 81-85.
Nagao, S. and M. Takahashi, 1954. Some genes responsible for yellow, brown and black color of glume. Genetical studies on rice plant, XVI. Japan. J. Breed. 4: 25-30.
Nagao, S. and M. Takahashi, 1963. Trial construction of twelve linkage groups in Japanese rice. Genetical studies on rice plant, XXVIII. J. Fac. Agr. Hokkaido Univ. 53: 72-130.
Takahashi, M., T. Kinoshita and K. Takeda, 1968. Character expression and causal genes of some mutants in rice plant. Genetical studies on rice plant, XXXIII. J. Fac. Agr. Hokkaido Univ. 55: 496-512.
2. Gene symbols for dwarfness
Yuzo Futsuhara, Faculty of Agriculture, Nagoya University, Chikusa-ku, Nagoya, 464 Japan
Fumio Kikuchi, Institute of Agriculture and Forestry, University of Tsukuba, Sakura-mura, Ibaraki, 305 Japan
J. Neil Rutger, USDA-ARS, Agronomy Department, University of California, Davis, CA 95616, USA.
A large number of dwarf mutants of spontaneous origin or induced by mutagen
treatments have been reported.
About 50 dwarfing genes (d-1 to d-50) were listed by Takahashi and Kinoshita (1977). Kinoshita and Shinbashi (1982) reported that the three relations, d-8, d-11 and d-14, d-10, d-15 and d-16, d-18h and d-18k are allelic respectively. Keeping these findings in mind 41 dwarfing genes are listed as shown in Table 1 (List of genes for dwarfness). Most of them are governed by a single recessive gene, with the exception of a dominant gene, D-53. Recently, Futsuhara and Kitano (1977) reported that most extreme dwarf mutants were also controllled by single recessive genes, but the mutants with slightly shortened stature, were largely determined by a single gene with imcomplete dominance.
Kada (1937) and Anonymous (1963) showed that the five recessive dwarfing genes designated d-1 to d-5 were nonallelic to each other. Reddy and Padma (1976) reported that five mutants of Tellakattera induced by ethylmethansulphonate treatment, were monogenic recessive. These were designated d-6 to d-10. Accordingly, it is necessary to clarify allelic relations among the dwarf genes by using materials from all over the world.
Foster and Rutger (1978) and Mackill and Rutger (1979) reported that a semidwarf gene, sd-1, induced from Calrose by r-ray irradiation, was allelic to the recessive dwarfing gene of Dee-geo-woo-gen (DGWG). In addition, Mackill and Rutger (1979) reported three nonallelic semidwarfing genes, sd-2, sd-3 and sd- 4. Thus two types of gene symbols, d and sd, have been used for the various degrees of dwarfness. However, the mutants described as sd and d are difficult to distinguish from each other and their expression is influenced by genetic backgrounds. It is therefore necessary that we adopt one of the two symbols (d or sd) for designating dwarfing mutants. However, some workers are in favor of continued use of d and sd. Therefore, further consultations are necessary to resolve this issue.
It is noteworthy that most of the semidwarfs of practical value in rice breeding have allelic dwarfing genes at sd-1 locus irrespective of their source of origin (Kikuchi and Ikehashi 1983). Thus useful dwarfing genes are limited to only one. It is terefore important to identify additional semidwarf genes non allelic to sd-1.
Table 1. List of genes for dwarfness
=============================================================================
Gene Name Linkage Chromo- Remark
group some
=============================================================================
d-1 daikoku dwarf VI 2
d-2 ebisu dwarf II 11
d-3 II 11 Triplicate genes
d-4 bunketsu-waito I 6 Triplicate genes
d-5 (tillering dwarf) X 8 Triplicate genes
d-6 ebisumochi dwarf or IV 10 Equivalent with d-34
tankan-shirasasa dwarf
d-7 heiei-daikoku or IV 10
cleistogamous dwarf
d-9 chinese dwarf I 6
d-10 kikeibanshinriki or III 3 Equivalent with
toyohikari bunwai d-15, d-16
(tillering dwarf)
d-11 shinkane-aikoku or II 11 Equivalent with
norin-28 dwarf d-8, d-14
d-12 yukara dwarf
d-13 short grained dwarf
d-14 kamikawa bunwai IX 5
(tillering dwarf)
d-17(t) slender dwarf
d-18h hosetsu-waisei or III 3 +>d-18k>d-18h
akibare dwarf (multiple alleles)
d-18k kotaketamanishiki dwarf
d-19 (t) kamikawa dwarf
d-20 hayayuki dwarf XII
d-21 aomorimochi-14 dwarf I 6
d-22 jokei 6549 dwarf
d-23 (t) ah-7 dwarf
d-24 (t) m-7 dwarf
d-26 (t) 7237 dwarf III 3
d-27 bunketsuto VIII 9
(tillering dwarf)
d-28 chokeidaikoku or VIII 9
long stemmed dwarf
d-29 short uppermost X 8
internode dwarf
d-30 waisei-shirasasa dwarf X 8
d-31 taichung-155 irradiated dwarf II 11
d-32 dwarf Kyushu-4 X 8
d-33 bonsaito dwarf 4
d-35 (t) tanginbozu dwarf
d-42 (t) liguleless dwarf II 11
sd-1=d-47(t)dee-geo-woo-gen dwarf III 3
d-49 (t) reimei type dwarf
d-50 (t) fukei 71 type dwarf
d-51 dwarf Kyushu-8 12
d-52 kwarf Kyushu-2 XI 5
D-53 Dwarf Kyushu-3 VIII 9
d-54 dwarf Kyushu-5 III 3
d-55 dwarf Kyushu-6 III 3
d-56 dwarf Kyushu-7 XI 5
d-57 dwarf VII 1
=============================================================================
(t), tentative symbol pending a futher test of allelism.
References
Anonymous 1963. Rice gene symbolization and linkage groups. U.S. Dep. Agr. ARS 34-28: 1-15.
Foster, K.W. and J.N. Rutger 1978. Independent segregation of semidwarfing genes and a gene for pubescence in rice. J. Hered. 69: 137-138.
Futsuhara, Y. and H. Kitano 1977. Genetical studies on short culm mutants in rice. IV. A diallel cross analysis of culm length. Japan. J. Breed. 27 (Supl. 2): 156-157. (in Japanese).
Kadam, B.S. 1937. Genes for dwarfing in rice. Nature 139: 1070.
Kikuchi, F. and H. Ikehashi 1983. Semidwarfing genes of high-yielding rice varieties in Japan. Gamma Field Symposia 22: 17-30.
Kinoshita, T. and N. Shinbashi 1982. Identification of dwarf genes and their character expression in the isogenic background. Japan. J. Breed. 32: 219-231.
Mackill, D.J. and J.N. Rutger 1979. The inheritance of induced-mutant semidwarfing genes in rice. J. Hered. 70:335-341.
Reddy, G.M. and A. Padma 1976. Some induced dwarfing genes non-allelic to Dee- Geo-Woo-Gen gene in rice, variety Tellakattera. Theo. Appl. Genet. 47: 115-118.
Takahashi, M. and T. Kinoshita 1977. List of genes and chromosome map of rice. In Plant Genetics IV. Morphogenesis and Mutation: 416-441. Syokabo, Tokyo. (in Japanese)
3. Gene symbols for heading behavior
Shigetoshi Sato, College of Agriculture, University of Ryukyus, Nishimachi, Okinawa-ken, 903-01 Japan
There are three series of gene symbols, E, Ef and Se, which have been assigned
to mutants controlling heading time and were listed in RGN 1. It is difficult
to conduct allelic tests between genes controlling heading time, since
continuous variation is usually observed in F\2\ populations of the crosses
between lines differing in days to heading. Isogenic lines are necessary for
the allelic tests. Isogenic lines of Taichung 65 with one or two genes for
earliness have been produced by Tsai (1976). Drs. Tsai and Sato have conducted
allelism tests between genes for basic vegetative growth. The tests showed that
the two genes for earliness, Ef-1 and Ef-2, were allelic (Tsai 1985a; Sato and
Nakasone 1985). Linkage analysis revealed that the Ef-1 gene is located on fgl-
linkage group (Nishimura's 7th chromosome) and the gene order is pgl-Rf-1-fgl-
Ef-1 (Sato et al. 1985; Sato and Shirakawa 1986).
At the Ef-1 locus, in addition to a recessive allele carried by Taichung 65, a series of dominant isoalleles have been detected, i.e., Ef-1a, Ef-1b, Ef- 1X, Ef-1r, etc., which showed significant differences in effects. These symbols are abbreviated as Ea, Eb, etc. in Tsai's publications. Furthermore, Tsai (1976, 1985a,b, 1986) has detected three more gene loci, i.e., m-Ef-1 (a recessive modifier emphasizing the effect of Ef-1, on Nishimura's 10th chromosome), lf-1(t) (a recessive gene delaying heading time, obtained from an induced mutant of Taichung 65, independent of the above loci, the linkage relations remain unknown), and lf-2(t) a recessive gene for late heading, found in a tropical Japonica variety, independent of all the above loci, but the locus is unknown. The lf-1(t) gene was found to render its carrier sensitive to photoperiod. No such effect was found for lf-2(t).
Three independent dominant genes delaying heading time, E\1\, E\2\ and E\3\ were detected at Kyoto University. These genes were found to control photoperiod senstivity (Yamagata 1984). E\1\ was independent of the photoperiod sensitivity gene, Se-1(Lm) (Okumoto et al. 1984). Se-1(Lm) belonged to linkage group I (chromosome 6), and was closely linked with a gene for blast-disease resistance, Pi-zt (Yokoo and Fujimaki 1971; Yokoo et al. 1980). It is plausible that Se-1 (Lm) is identical or allelic to other photoperiod sensitivity genes, fl (Jodon 1940), Se (Chandraratna 1953), and Rs (Kudo 1968). In addition, a recessive gene, se-2 belonging to linkage group IV is mentioned by Yu and Yao (1968).
References
Chandraratna, M.F., 1953. A gene for photoperiod sensitivity in rice linked with apiculus color. Nature 171: 1162.
Jodon, N.E. 1940. Inheritance and linkage relationship of chlorophyll mutation in rice. J. Amer. Soc. Agron. 32: 342-346.
Kinoshita, T., 1984. Current linkage maps. List of genes and genetic stocks. RGN 1: 16-77.
Kudo, M. 1968. Genetical and breeding studies on physiological and ecological characters in hybrids between ecological groups of rice. Bull. Nat. Inst. Agr. Sci. Ser. D19:1-84. (In Japanese with English summary).
Okumoto, T., T. Tanisaka and H. Yamagata, 1984. Genetical analysis for heading trait of rice varieites in Japan. 7. Linkage relationship between heading-date gene E\1\ and blast-resistance gene Pi-zt. Japan J. Breed. 34 (Suppl. 1): 292-293. (In Japanese).
Sato, S. and I. Sakamoto, 1983. Inheritance of heading time in isogenic line rice cultivar, Taichung 65 carrying earliness gene from a reciprocal translocation homozygote, T3-7. Japan. J. Breed. 33 (Suppl.1):118-119. (In Japanese).
Sato, S., I. Sakamoto and S. Nakasone, 1985. Location of Ef\1\ for earliness on Nishimura's seventh chromosome. RGN 2:59-60.
Sato, S., and K. Shirakawa, 1986. Determination of an earliness gene locus on Nishimura's seventh chromosome of rice Oryza sativa L. Japan. J. Breed. 36(Suppl.1):354-355. (In Japanese).
Tsai, K.H., 1976. Studies on earliness genes in rice, with special reference to analysis of isoalleles at the E locus. Jpn. J. Genet. 51: 115-128.
Tsai, K.H., 1985a. Further observations on the Ef-1 gene for early heading. RGN2: 77-78.
Tsai, K.H., 1985b. Analysis of genes for late heading in rice. In New Frontiers of Breeding Research. Proc. 5th Intern. Congr. SABRAO, p. 221-236. Kasetsart University, Bangkok.
Tsai, K.H., 1986. Gene loci and alleles controlling the duration of basic vegetative growth of rice. In Rice Genetics, Proc. Intern. Rice Genet. Symp. May 1985., p. 339-349. IRRI publication.
Yamagata, H., 1984. Heading time genes of rice, E\1\, E\2\ and E\3\. RGN 1: 100-101.
Yokoo, M. and H. Fujimaki, 1971. Tight linkage of blast-resistance with late maturity observed in different Indica varieties of rice. Japan. J. Breed. 21: 35-39.
Yokoo, M., F. Kikuchi, A. Nakane and H. Fujimaki, 1980. Genetical analysis of heading date by aid of close linkage with blast resistance in rice. Bull. Nat. Inst. Agric. Sci. D 31: 95-126.
Yu, C.J. and T.T. Yao, 1968. Genetische Studien beim Reis. II. Die Koppelung des Langhullspelzengens mit dem Photoperiodizitatsgen. Bot. Bull. Acad. sinica 9: 34-35.
4. Gene symbols for anthocyanin coloration
Toshiro Kinoshita, Plant Breeding Institute, Faculty of Agriculture, Hokkaido University, Sapporo, 060, Japan.
Marker genes responsible for anthocyanin pigments are important for chromosome
mapping. A wide range of variations exists in the intensity and hues of
coloration both in Japonica and Indica varieties. It has been postulated that
the three loci, C, A and P, are responsible for the various anthocyanin
pigments which are expressed principally in the apiculus coloration.
According to Nagao, Takahashi and their co-workers, the intensity of
pigmentation is due to the interaction of the multiple alleles at the three
loci. The order of dominance of alleles at these loci is:
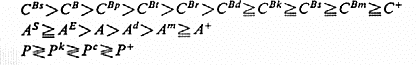
Setty and Misro (1973) confirmed this scheme in Indica strains and suggested
that any two of genes, Pa, Pb and Pc, together acted as basic genes
instead of alleles at P locus.
As to the pigmentation of plant parts other than the apiculus, various kinds of distributing genes are interacting with inhibitors and anti-inhibitors. Genes postulated by Japanese and Indian workers are shown in Table 1 (Genes postulated for anthocyanin colorations in Japonica and Indica rice).
It is difficult to determine the identity of the genes from the literature because the expression of color shades and hues are greatly modified by the environmental conditions and genetic backgrounds. There is a possibility of some discrepancy in gene identification even if the same gene symbol is assigned to a certain pigmentation. Allelic tests are needed to confirm the particular distributing gene under the co-existence of higher rank alleles of the basic three genes. Hsieh and Chang (1964) postulated that chromogen gene Prp-a (Pp) acts complementarily with Prp-b(Pb) for purple pericarp, while Nagao et al. (1962) explained the same phenomena by the pleiotropic effect of an allele, PlW at the Pl-locus in co-existence with C-A-P genes. In another case, the expression of purple pericarp was reported to be controlled by two pairs of duplicate genes (Rao and Seetharaman 1973).
Table 1. Genes postulated by anthocyanin colorations in Japonica and Indica
rice
=============================================================================
Character Japonica Indica
=============================================================================
Coleoptile Pleiotropic action of basic Pc\1\(III),Pc\2\,I-Pc,Ai-Pc(IV)
genes, C, A and P
Apiculus C(I),A(III),P(II) Pa(IV),I-P,Ai-P(IV),Ap(IV)
Lemma Pr(II) Pr(X),Pr\2\,Pr\3\,A,Pr\a\(IV)
Glume(empty) Pleiotropic action of basic Pg\1\(X),Pg\2\(III),Pg\3\,Ai-Pg(II)
genes, C, A and P A,Pg(IV),Gp,I-Gp(III)
Stigma Ps-1(V),Ps-2(II) or Ps-3(II) A,Ps\a1\,Ps\a2\,Ps\a3\,Ps(III),I-Ps
Leaf blade Pl, Plw,Pli(II)* C,A,Lsp\1\,Lsp\2\,Ilp etc.
Leaf axil Px(III)
Leaf sheath Pl,I-Pl or Plw I-Pl** C,A,Lsc,Psh(III)
Auricle Pleiotropic action of Plw Pau\a\(III),Pau\b\,I-Pau,Ai-Pau(IV)
Ligule Pleiotropic action of Pl, Plg\a\(X),Plg\b\(X),Ai-Plg(IV)
Plw or Pli
Junctura Pn(III),Pleiotropic action Pj\a\(V),Pj\b\,Pj\c\,Pj\d\,Pj\e\,
of Pl Pj\a\(III),Pj\b1\,Pj\b2\,I-Pj
Nodal ring Pleiotropic action of Plw Pnr\1\(X),Pnr\2\,Pnr\3\
Node Pn(III) Pn\1\(X),Pn\2\,Pn\3\,Pn(III)
Pulvinus Pn(III) Pu\a\(III),Pu\b\,Pu\c\,Pu\d\
Septum Pm\a\(III),Pm\b\,Pm\c\,Pm\d\
Internode Pin-1(II) or Plw,I-Pl Pin\a\(III),A,Pin\a\(IV),Pin\a1\(IV)
=============================================================================
* Multiple alleles
References
Hsieh, S.C. and T.M. Chang, 1964. Genic analysis in rice. IV. Genes for purple pericarp and other characters. Japan J. Breed. 14: 141-149.
Kinoshita, T., 1984. Gene analysis and linkage map. In Biology of Rice. eds by S. Tsunoda and N. Takahashi: 187-274. Japan Sci. Soc. Press Tokyo/Elsevier, Amsterdam.
Nagao, S., M. Takahashi and T. Kinoshita, 1962. Genetical studies on rice plant. XXVI. Mode of inheritance and causal genes for one type of anthocyanin color character in foreign rice varieties. J. Fac. Agr. Hokkaido Univ. 52: 20- 50.
Rao, C.H. and R. Seetharaman, 1973. Genetic studies in pericarp and hull colour in rice. Indian J. Genet. 33: 319-323.
Setty, M.V.N. and B. Misro, 1973. Complementary genic complex for anthocyanin pigmentation in the apiculus of rice (Oryza sativa L.). Can. J. Genet. Cytol. 15: 779-789.
Recent publications on the inheritance studies of anthocyanin coloration
Das, G.R. and T. Ahmed, 1982. Distribution of pigments in glutinous rice. Oryza
19: 114-116.
Eruotor, P.G., 1986. Inheritance of sheath colouration and of brown leaf spot of rice. Z. Pflanzenzuchtg. 96: 47-52.
Goud, J.V. and B.Y. Kullaiswamy, 1984. Pleiotropy and differential action of genes in rice (Oryza sativa L.). Indian J. Genet. 44: 262-265.
Hadagal, B.N., A. Manjunath and J.V. Goud, 1984. Inheritance of anthocyanin pigmentation in a few parts of rice (Oryza sativa L.). Indian J. Genet. 44: 319-324.
Kinoshita, T. and M. Maekawa, 1986. Inheritance of purple leaf color found in Indica rice. -Genetical studies on rice plant, SCIV-. J. Fac. Agr. Hokkaido Univ., 62: 453-466.
Kolhe, G.L. and N.R. Bhat, 1982. Linkage of five anthocyanin genes in rice. Indian J. Genet. 42: 92-100.
Maekawa, M. and F. Kita, 1983. Spectral properties of extracted pigments from genic colored grains of rice. Res. Bull. Univ. Farm, Hokkaido Univ. 23: 11-21. (In Japanese with English summary).
Ramesh, B., 1984. Inheritance of anthocyanin pigment in rice. Indian J. Genet. 44: 15-17.
Tripathi, R.S. and M.J. Balakrishna Rao, 1984. Inheritance of photoperiod sensitivity and apiculus pigmentation in rice. Indian J. Agric. Sci. 54: 157- 164.
Yadav, L.N., 1984. Inheritance of anthocyanin pigmentation in stigma, outer glumes and apiculus of rice (Oryza sativa L.)-I.J. Indian Bot. Soc. 63: 186- 189.
Yadav, L.N., and N.S. Tomar, 1984. Inheritance of anthocyanin pigmentation in stigma, outer glumes and apiculus of rice (Oryza sativa L.). II. J. Indian Bot. Soc. 63: 190-193.
5. Gene symbols for grain size and shape
Kazuyoshi Takeda, Okayama University, Kurashiki, Okayama-ken, 710 Japan
To date only a few major genes controlling the grain size and shape have been
reported. Grain size may be indicated by weight, volume, length, etc., but
weight and length are more commonly used as indicators of grain size. Grain
length closely correlates with grain weight. When grain shape is expressed as
length/breath ratio, grain length may also correlate with grain shape index.
Table 1 (Variation and correlation of size and shape of rice kernel (brown
rice) in 88 varieties with wide range of variation), shows an example of the
above-mentioned relationship between the components of grain size and shape.
These statistics are deduced from a sample of 88 rice varieties with a very
wide range of variation in grain size and shape (Fig.1 Variation in size and
shape of rice kernels.)
Long kernels are usually large in size and slender in shape. In other words, the gene or genes which determine kernel length may also determine size (weight) and shape of the kernel. However, the gene designation may be based on the most impressive feature, such as size, length or shape of the grain. The gene symbols used for grain characters to date are; bk (for big kernel), Mi (for minute kernel), Lk (for long kernel), rk (for round kernel) (cf. RGN 1, p. 34-35). I propose the gene symbols for size and shape of rice grain as listed in Table 2 (gene symbols for size and shape of rice grain). In case a new gene for short kernel is later found to be allelic to long kernel gene Lk, the multiple alleles at this locus may be designated as Lk (long), Lk+ (normal) and LkS (short), and so on.
Recently, Tomer (1985) reported a multiple allelic series Gl\1\, Gl\2\ and gl for grain length. However, these gene symbols are unacceptable because: (1) gl has been allotted for glabrous character, (2) multiple alleles should be designated by superscript, and (3) his allelic tests seem to be incomplete.
Table 1. Variation and correlation of size and shape of rice kernel (brown
rice) in 88 varieties with wide range of variation
Fig. 1. Variation in size and shape of rice kernels.
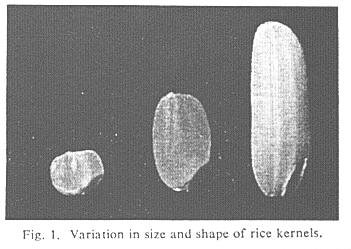
Table 2. Gene symbols for size and shape of rice grain
-----------------------------------------------------------
Character Gene symbol
-----------------------------------------------------------
Kernel size Lk for long kernel
Shk for short kernel
-------------------------
Bk for big kernel
Mi for minute kernel
-----------------------------------------------------------
Kernel shape Slk for slender kernel
Rk for round kernel
-----------------------------------------------------------
Tomer, J.B., 1985. Studies on the inheritance of kernel size and its association with physical and chemical quality characters in rice (Oryza sativa L.). Z. Pflanzenzuchtg. 95: 361-366.
6. Gene symbols for isozymes
Hiroko Morishima, National Institute of Genetics, Yata, Mishima, Shizuoka-ken, 411 Japan and J.C. Glaszmann, International Rice Research Institute, P.O. Box 933, Manila, Philippines
More than 40 loci encoding isozymes have been proposed in rice, although only a
few of them have been located on respective chromosomes. Various techniques
have been used to detect isozymes. Consequently, it is difficult to determine
the correspondance between the results of different studies. This has resulted
in the use of different gene symbols. To prevent further confusion, we propose
the use of a unified system of gene designation as follows:
(1) Three letters (beginning with a capital) to denote the name of the enzyme.
(2) One numeral to identify a particular locus among those which encode the various molecular forms of the same enzyme.
Table 1 (List of genes for isozymes), summarizes the relationships between the existing gene symbols and proposed new designations following the above rules. For the convenience of isozyme workers and other researchers, all loci for which Mendelian segregations have been confirmed are listed in the table. Some of these have been published but others are from personal communications. Loci have been numbered according to precedence of publication and order of band mobility.
Various systems have been adopted for designating alleles at the same locus. Use of a common nomenclature requires the establishment of a set of marker varieties in a Rice Genetic Stock Center. We propose the use of a numeral for alleles represented in the Stock Center and use of letters for alleles not tested against the available markers.
Table 1. List of genes for isozymes
=============================================================================
Proposed Enzyme Published synonyms (ref.)
locus =================================================
symbol (E.C. No) (1) (2) (3) (4)
=============================================================================
Acp-1 Acid phosphatase Acp-B(Pac-AMC) Acp-1
Acp-2 (E.C.3.1.3.2) Acp-C(Pac-F-S) Pac-2 Acp-2
Acp-3 Acp-3
Acp-4 Pac-1
=============================================================================
Adh-1 Alcohol
dehydrogenase Adh-A
(E.C.1.1.1.1)
=============================================================================
Amp-1 Aminopeptidase Lap-E Lap
Amp-2 (E.C.3.4.11.-) Aap Amp-1
Amp-3 Amp-2
=============================================================================
Cat-1 Catalase Cat-A Cat Cat-1
=============================================================================
Est-1 Esterase Est-D Est-3 Est-1
Est-2 (E.C.3.1.1.-) Est-E Est-4 Est-2 Est-2
Est-3 Est-J Est-3
Est-4 Est-4
Est-5 Est-B
Est-6 Est-C Est-2
Est-7 Est-I Est-5
Est-8 Est-6
Est-9 Est-Ca Est-1 Est-c1
=============================================================================
Got-1 Aspartate
aminotranspherase Got-A
Got-2 (E.C.2.6.1.1.) Got-B
Got-3 Got-C
=============================================================================
Icd-1 Isocitrate
dehydrogenase Icd-A(Idh-A)
(E.C.1.1.1.42)
=============================================================================
Mdh-1 Malate
dehydrogenase Mdh-A
(E.C.1.1.1.37)
=============================================================================
Pgd-1 Phosphogluconate Pgd-A Pgd-1
Pgd-2 dehydrogenase Pgd-B
(E.C.1.1.1.43)
=============================================================================
Pgi-1 Phosphoglucose Pgi-A Pgi-1 Pgi-1
Pgi-2 (E.C.5.3.1.9) Pgi-B Pgi-2 Pgi-2
Pgi-3 Pgi-3
=============================================================================
Pox-1 Peroxidase Pox-E Px-1
Pox-2 (E.C.5.3.1.9) Px-2
Pox-3 Pox-B Px-3
Pox-4 Pox-C
=============================================================================
Sdh-1 Shikimate
dehydrogenase Sdh-1
(E.C.1.1.1.25)
=============================================================================
(1) Second, G. and P. Trouslot, 1980. Electrophorese d'enzymes de riz (Oryza sp.). Travaux et Documents de l'ORSTOM, No. 120, 88p.
Second, G., 1982. Origin of the genic diversity of cultivated rice (Oryza spp.): Study of the polymorphism scored at 40 isozyme loci. Jpn. J. Genet. 57: 25-57.
Second, G., 1985. Evolutionary relationships in the Sativa group of Oryza based on isozyme data. Genetique, Selection, Evolution, 17(1):89-114.
(2) Glaszmann, J.C., H. Benoit and M. Arnaud, 1984. Classification de riz cultives (Oryza sativa L.): Utilisation de la variabilite isoenzymatique. Agr. Trop. 39(1):51-66.
Glaszmann, J.C., 1986. A varietal classification of Asian cultivated rice (Oryza sativa L.) based on isozyme polymorphism. In Rice Genetics, 83-90. IRRI Publication.
(3) Pai, C., T. Endo and H.I. Oka, 1983. Genic analysis for peroxidase isozymes and their organ specificity in Oryza perennis and O. sativa. Can. J. Genet. & Cytol. 15: 845-853.
Pai, C., T., Endo and H.I. Oka, 1975. Genic analysis for acid phosphatase in Oryza perennis and O. sativa. Can. J. Genet & Cytol. 17: 637-650.
Pai, C. and P.Y. Fu, 1977. genetic analysis for peroxidase and acid phosphatase isozymes in cultivated rice. Agric. Bull, Taiwan, 2: 75-85.
Morishima, H. and R. Sano, 1984. Genic analysis for isozymes in rice. RGN 1: 117-118.
Sano, R. and P. Barbier, 1985. Analysis of five isozymes genes and chromosomal location of Amp-1. RGN 2: 60-62.
(4) Nakagahra, M., 1977. Genic analysis for esterase isozymes in rice cultivars. Japan. J. Breed. 27: 141-148.
Nakagahra, M., 1978. The differentiation, classification and center of genetic diversity of cultivated rice by isozyme analysis. Trop. Agric. Res. Ser. 11: 77-82.
IV. List of gene symbols
Gene symbols newsly adopted and linkage relations newly confirmed are listed.These are supplements to the lists presented in RGN 1 and 2. A comprehensive list of gene symbols and linkage groups will be presented in a future issue of RGN after completion of the work on revision of gene symbols and adoption of a unified system of chromosome numbering.
a. List of gene symbols (Supplement)
============================================================================= Acp-4 (Acp-A) Acid phosphatase-4 (E.C. 3.1.3.2) Adh-1 (Adh-A) Alcohol dehydrogenase (E.C. 1.1.1.1) Cts-1 Cold tolerance at seedling stage d-58(t) small grained dwarf Est-5 (Est-B) Esterase-5 (E.C. 3.1.1.-) Est-6 (Est-C) Esterase-6 (E.C. 3.1.1.-) Est-7 (Est-I) Esterase-7 (E.C. 3.1.1.-) Est-8 Esterase-8 (E.C. 3.1.1.-) Got-1 (Got-A) Aspartate aminotranspherase-1 (E.C. 2.6.1.1) Got-2 (Got-B) Aspartate aminotranspherase-2 (E.C. 2.6.1.1) Got-3 (Got-C) Aspartate aminotranspherase-3 (E.C. 2.6.1.1) Icd-1 (Icd-A, Idh-A) Isocitrate dehydrogenase-1 (E.C. 1.1.1.42) Nd Early nodal differentiation Nr-1 Nodal rooting Nr-2 (complementary genes) Pgd-2 (Pgd-B) Phosphogluconate dehydrogenase-2 (E.C. 1.1.1.43) Pgi-3 Phosphoglucose isomerase-3 (E.C. 5.3.1.9) Pi-sh Pyricularia oryzae resistance-sh Pox-4 (Pox-C) Peroxidase-4 (E.C. 1.11.1.7) Shp-1 Sheathed panicle-1 shp-2 sheathed panicle-2 =============================================================================
============================================================================= Old New Name ============================================================================= Amp-1 Amp-3 Aminopeptidase-3 (E.C. 3.4.11.-) Ef-2 Ef-1 Earliness-1 Est-c1 Est-9 Esterase-9 (E.C. 3.1.1.-) fes-1 fes-1, fes-2 female sterility-1, -2. Fes-2 Fes-3 Female sterility-3 Lap-1 Amp-1 Aminopeptidase-1 (E.C. 3.4.11.-) =============================================================================
=============================================================================
Gene Name Gene locus Reference
=============================================================================
Group III (A group)
Shp-1 Sheathed panicle-1 71 4
Group VI+IX (d-1 group)
bgl bright green leaf 79 2
ri verticillate rachis 82 2
spl-7 spotted leaf-7 97 2
nl-1 neck leaf-1 107 2
al-2 albino-2 108 2
=============================================================================
Cytological map of fgl group
Reference
pgl-7-8b-Rf-1-fgl-7-11-Ef-1-6-7-3-7-7-9-7-8a 6
=============================================================================