|
Although grain size in cereal crops is under polygenic control, several
major genes for this trait have been identified in rice. One such gene,
Minute (Mi), was identified in the central region of rice
chromosome 3, and found to impart a small-grain phenotype in an incompletely
dominant manner (Takeda and Saito, 1977). Mi and other major genes
have proved to be useful markers in morphological mapping studies. In
this study, we mapped Mi using microsatellite (SSR) markers to
place it more precisely on the high-density molecular map of rice.
A line of rice homozygous for Mi, H-343, was crossed with the rice
variety IR64 to develop a segregating F2 population of 113
individuals that was used for mapping. These individuals were evaluated
for six different phenotypes: seed length (SL), grain length (GL), seed
weight (SWe), grain weight (GWe), seed width (SWi) and grain width (GWi).
All individuals were also genotyped using previously published SSR markers
on chromosome 3 (Li et al., 2004 and Temnykh et al., 2001).
Using the computer software Mapmaker V2.0 (Lander et al., 1987),
an integrated linkage map of chromosome 3 was created and interval analysis
was carried out using the Q-gene software package (Nelson, 1997). Empirical
significance thresholds for declaring a QTL were similar for all traits
evaluated, with an average threshold value of LOD>2.95 at the p<0.01
significance level.
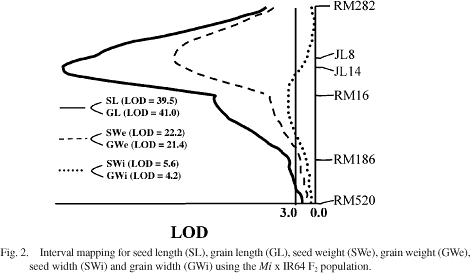
Results of interval mapping for the traits seed length (SL), grain length
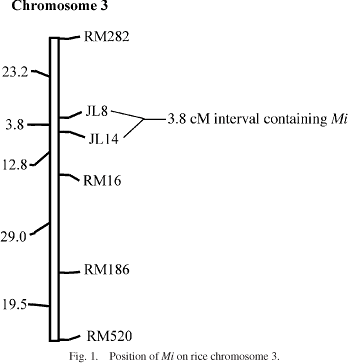
(GL), seed weight (SWe) and grain weight (GWe) positioned Mi in
the interval between markers RM282 and RM16, with peak LOD scores occurring
in or within 2 cM of the 3.8 cM interval between markers JL8 and JL14
(Figure 1). Peak LOD scores for SL, GL, SWe and GWe were 39.5, 41.0, 22.2
and 21.4 respectively (Figure 2). Interval mapping for the traits seed
width (SWi) and grain width (GWi) indicated that the locus associated
with these phenotypes differed from the Mi locus reported above.
The peak LOD scores for these traits were 5.6 for SWi and 4.2 for GWi,
with both occurring in the interval between markers JL8 and RM186, and
falling closer to RM16 than to either of its flanking markers (Figure
2).
These results suggest that there are two separate genes on chromosome
3 that affect grain size: one gene, which is believed to be Mi,
has a large effect on grain length and weight, while the other, linked
gene has a smaller effect on grain width. These findings are consistent
with the conclusion of Kato (1989) that grain length and grain width are
generally controlled by different genetic systems. Furthermore, the low
LOD scores obtained at the width locus are consistent with results generally
seen for polygenes, where any one locus explains only a small portion
of the phenotypic variation associated with a quantitative trait. This
is in contrast to the much higher LOD scores achieved at the putative
Mi locus, which are consistent with those associated with genes
exhibiting major effects on a phenotype. Previous work done on Mi
also suggests that this gene is most closely associated with the phenotype
grain length (Takeda and Saito, 1977). Thus, results of the interval mapping
completed in this study indicate that Mi is most likely located
in the 3.8 cM interval between SSR markers JL8 and JL14, both located
in the genetically defined centromere region of chromosome 3.
Acknowledgments
We gratefully acknowledge Dr. Itsuro Takamure from Hokkaido University
who provided the original Minute seeds and Dr. Neil Rutger at the
Dale Bumpers National Research Center in Stuttgart, Arkansas, USA for
help with phenotypic evaluation of seeds.
References
Kato, T., 1989. Diallel analysis of grain size of rice (Oryza sativa
L.). Japan. J. Breed. 39: 39-45.
Lander, F., P. Green and L. Abrahamson, et al. 1987. Mapmaker:
an interactive computer package for constructing primary genetic linkage
maps of experimental and natural populations. Genomics 1: 174-181.
Li. J., M. Thomson and S. McCouch, 2004. Fine mapping of a grain weight
QTL in the pericentromeric region of rice chromosome 3. Genetics: in press.
Nelson, J.C., 1997. Q-gene: software for marker-based genomic analysis
and breeding. Mol. Breed. 3: 239-245.
Takeda, K. and K. Saito, 1977. The inheritance and character expression
of the minute gene derived from a rice genetic tester Minute. Bull.
Fac. Agr. Hirosaki Univ. 27: 1-29. (in Japanese with English summary)
Temnykh, S., G. DeClerck and A. Lukashova, et al. 2001. Computational
and experimental analysis of microsatellites in rice (Oryza sativa
L.): frequency, length variation, tansposon associations, and genetic
marker potential. Genome Res. 11: 1441-1452.
|